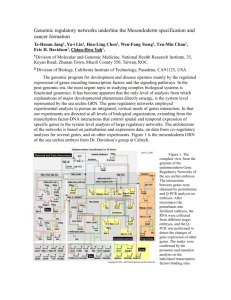
developmental biology of the sea urchin xxii
advertisement

DEVELOPMENTAL BIOLOGY OF THE SEA URCHIN XXII APRIL 23 – 27, 2014 MARINE BIOLOGICAL LABORATORY WOODS HOLE, MA The sea urchin featured on the front cover is Coelopleurus maculatus, Family Arbaciidae. It was collected by fisherman's net from 100m depth in the entrance area of Tokyo Bay and given to Dr. Masato Kiyomoto, Director of the Tateyama Marine Laboratory. DEVELOPMENTAL BIOLOGY OF THE SEA URCHIN XXII APRIL 23 – 27, 2014 MARINE BIOLOGICAL LABORATORY WOODS HOLE, MA Oganizers: Dave McClay Eric Davidson David Burgess Julia Morales The Organizers would like also to acknowledge the generous support of the following sponsors: The Society for Developmental Biology lumencor TABLE OF CONTENTS Program 2-6 Poster Presenters 7-8 Abstracts. Talk Sessions 9 – 36 Abstracts, Posters 37 - 51 Participants List 52 PROGRAM Sea Urchin XXII Wednesday April 23 A Dinner 6:00-7:30 pm Evening Opening Plenary Session: Complex Spatial patterning following gastrulation 7:30-9:00 PM Lillie Auditorium Chair, Dave McClay 7:30 Isabelle Peter: Use of regulatory complexity in gut organogenesis 8:00 Dave McClay: Spatial regulatory states in the ectoderm. 8:30 Thierry Lepage: A maternal determinant of dorsal-ventral axis in Paracentrotus lividus. Posters/Mixer 9:00-11:00 pm Meigs Room Thursday April 24 Breakfast 7:00-8:30 am Plenary Session II: Genomic Logic and Cell Specification 8:30-12:00 Lillie Auditorium Chair, Robert Burke 8:30 Eric Davidson: Sea urchin GRN model 9:00 Patrick Lemaire: Similar embryos with divergent genomes: the ascidian paradigm 9:30 Robert Burke: Neurogenesis Coffee Break 10:00-10:30 Lillie Auditorium 10:30 Steve Small: Transcriptional networks and gradients that control body plan formation in Drosophila 11:00 Len Zon: Programs Regulating Stem Cell Self Renewal and Migration 11:30 Chuck Ettensohn: Morphogenesis of the Embryonic Skeleton Lunch 12:00-1:30 pm Plenary session III: Information at the bottom of the egg 1:30-3:00 PM Lillie Auditorium Chair, Julia Morales 1:30 Athula Wikramanayake: Deconstructing the Animal-Vegetal Axis 2:00 Amro Hamdoun: Membrane transport activity changes necessary for small micromere motility and pigment cell formation. 2:30 Zak Swartz: Post transcriptional mechanisms for germ line segregation in echinoderms Coffee Break 3:00-3:30 pm Lillie Auditorium Concurrent Session A: Gametes, Fertilization & Cleavage 3:30-5:45 PM Speck Auditorium Chair, Brad Shuster 3:30 Dominic Poccia: Quantification of Exocytosis Kinetics by DIC Image Analysis of Cortical Lawns 3:50 Vanessa Zazueta, Hedgehog signaling functions during sea star development 4:10 Nathalie Oulhen, Nanos protein enrichment in the small micromeres 4:30 Nadine Peyrieras: Sea urchins as models for the construction of prototypic embryonic development 4:50 Joe Campanale: ATP-binding cassette transporters mediate small micromere migration and left/right coelomic pouch segregation in Strongylocentrotus pupuratus 5:10 Henson, John: Inhibition of the Arp2/3 Complex in Sea Urchin Coelomocytes Induces a Lamellipodial to Filopodial Shape Change and Alters the Cell Spreading Process Concurrent Session B: Specification toward specific cell types 3:30-5:45 PM Lillie Auditorium Chair, Chuck Ettensohn, 3:30 Julius Barsi: General approach for in vivo recovery of cell-type specific effector gene sets 3:50 Ryan Range Wnt signaling modulators establish an anterior signaling center that patterns the anterior neuroectoderm territory in the sea urchin embryo 4:10 Jeni Croce: A Comprehensive Survey of Wnt and Frizzled Expression in the Sea Urchin Paracentrotus lividus 4:30 Oliver Krupke: Pigment cell migration: The role of Eph-Ephrin signaling 4:50 Megan Martik: Mechanisms of Small Micromere Homing 5:10 Catherine Schrankel: Transcriptional control of immune cell development in the purple sea urchin embryo Dinner 5:45-7:15 pm Plenary Session IV: New model systems for deuterostome development 7:30-9:00 pm Lillie Auditorium Chair, Veronica Hinman 7:30 Dan Medeiros: Insights from amphioxus and lamprey into the evolution of vertebrate head skeleton development 8:00 Paola Oliveri: Amphiura filiformis: an emerging model system to study skeletogenesis during development and regeneration 8:30 Chris Lowe: The role of Nodal signaling in axis formation of the hemichordate Saccoglossus kowalevskii Posters and Mixer 9:00-11:00 pm Meigs Room Friday April 25 Breakfast 7:00-8:30 am Plenary Sesson V. Specification 8:30am-12pm Lillie Auditorium Chair, Ina Arnone 8:30 Andy Ransick: Cis-reg logic of Ese and Prox genes underlying output in blastocoelar mesoderm founders 9:00 Yi-Hsien Su: Asymmetrical stabilization of hypoxia inducible factor α during sea urchin embryogenesis 9:30 Enhu Li - Gene Regulatory Network Governing 2-Dimensional Expression Patterns in the Sea Urchin Ectoderm Coffee Break 10-10:30 pm Lillie Auditorium 10:30 Miao Cui: A global survey of Wnt signaling in the early development of purple sea urchin embryos 11:00 Carmen Andrikou - A gene regulatory network orchestrating myogenesis in the sea urchin embryo 11:30 Cyndi Bradham: New Genes in Dorsal-Ventral Skeletal Patterning Lunch 12:00-1:30 pm Plenary Session VI: Motile components important for early divisions 1:30-3:00 PM Lillie Auditorium Chair, Gary Wessel 1:30 Brad Shuster: Dynamics and regulation in the early embryo 2:00 Alex McDougall: Three cell behaviors involved in shaping the ascidian gastrula : cell cycle duration, unequal cleavage, and oriented cell division. 2:30 David Burgess: Membrane domains regulating cleavage Coffee Break and Group Photo 3:00-3:30 pm Lillie Auditorium Concurrent Session C: Embryonic molecular biology 3:30-5:45 PM Speck Auditorium Chair, Jia Song 3:30 Jia Song: Functional analysis of microRNAs in development. 3:50 Smadar Ben-Tabou de-Leon: The ectoderm-mesoderm connection and the upstream regulation of VEGF and VEGFR. 4:10 Michael Piacentino: Alk4/5/7 Activity is Required for Animal Skeletal Patterning. 4:30 Dolores (Loli) Molina Jiménez: Segregation of pigment and blastocoelar cells depends on the interplay between the lineage specific transcription factors ESE and GCM 4:50 Zheng Wei: SoxC functions in neural precursor cells in sea urchin embryo neurogenesis. 5:10 Vincenzo Cavalieri: Suppression of nodal expression in prospective dorsal cells of the early sea urchin embryo by the Hbox12 homeodomain regulator. Concurrent Session D: Evolutionary Mechanism 3:30-5:45 PM Lillie Auditorium Chair, Yi-Hsien Su 3:30 Dede Lyons: Evolution of ectoderm-mesoderm communication during skeletal patterning in echinoid larva 3:50 Feng Gao Experimental approach to divergence in test organization between euechinoid and cidaroid sea urchins 4:10 Margherita Perillo: Evolution of pancreatic cell types 4:30 Jon Valencia: Regulatory gene use within the gastrulating sea urchin 4:50 Eric Erkenbrack: Delta-Notch signaling and HesC mediate the spatial confinement of the skeletogenic-specific regulatory gene alx1 to micromeredescendants in Eucidaris tribuloides 5:10 Alys Cheatle: Transcription factor evolution. Dinner 5:45-7:15 pm Plenary Session VII: Intracellular mechanisms that drive early development 7:30-9:00 Lillie Auditorium Chair, David Burgess 7:30 Gary Wessel: Something awesome about echinoderms 8:00 Michael Whitaker: Activation of DNA and protein synthesis at fertilization in the sea urchin. 8:30 Julia Morales: Translatome analysis following fertilization Posters and Mixer 9:00-11:00 pm Meigs Room Saturday April 26 Breakfast 7:00-8:30 A Plenary Session VIII: Evolution 8:30-12:00 Lillie Auditorium Chair, Greg Wray 8:30 Greg Wray: Intraspecific variation in GRNs 9:00 Jeff Thompson: A prelude to the present: Paleontological perspectives on 450 million years of echinoid evolution 9:30 Ina Arnone: Pattern and process during gut morphogenesis: an evolutionary perspective Coffee Break 10:00-10:30 am Lillie Auditorium 10:30 Jonathan Rast: Differentiation of immunocytes and the emergence of the larval immune system 11:00 Veronica Hinman: A gene regulatory network for neurogenenesis in development and regeneration 11:30 Stephan Schneider: Spiralian embryogenesis at your fingertips: the quest for an early annelid GRN Lunch 12:00-1:30 pm Plenary Session IX: Differentiation of cell types 1:30-3:00 Lillie Auditorium Chair, Jonathan Rast 1:30 Mamiko Yajima – Developmental plasticity: A broad utilization of germ line molecules in multipotent cells of the sea urchin 2:00 ShunsukeYaguchi: Maintenance of the anterior neuroectoderm of the sea urchin embryo 2:30 Courtney Smith: Coelomocytes and the Sea Urchin Immune System Coffee Break 3:00-3:30 pm Lillie Auditorium Concurrent Session E: Embryo transcriptomes and Genomics 3:30-5:00 PM Lillie Auditorium Chair: Andy Cameron 3:30 Jennifer Wygoda: Transcriptome analysis of direct vs indirect development 3:50 David Dylus: De-novo developmental transcriptome of the brittle star Amphiura filiformis 4:10 Andy Cameron: Sequencing Echinoderm Genomes 4:30 Sarah Tulin: Genome wide identification of regulatory elements 4:50 Jongmin Nam: Experimental measurement of embryonic regulatory states Concurrent Session F. Cytoskeleton and signaling. 3:30-5:00 PM Speck Auditorium Chair, Cyndi Bradham 3:30 Kathleen Moorhouse, cytoskeleton and polarity 3:50 Tara Fresques: Germ cell associated gene expression is regulated by Nodal signaling in the sea star P. miniata. 4:10 Bob Morris: Hedgehog and cilia 4:30 Andrea Bodnar Tissue homeostasis, regeneration and negligible senescence: insight from the sea urchin 4:50 Meike Stumpp: Deuterostome origins of gastric pH regulation Business Meeting: Lillie Auditorium 5:15 - 5:30 pm POSTER SESSION/Mixer: 5:30-7:00 pm Swope Hall BANQUET : 7:00 – 8:30 pm Sunday April 27 Breakfast 7:00-8:30 Check-Out 10:00 am - Pick up Bag Lunch Poster Presenters 71 The small GTPase Arf6 is essential for the development of the sea urchin larval gut Ahiakonu, Priscilla; Stepicheva, Nadezda; Dumas, Megan; Song, Jia L. 72 1-MA signaling and Hox cluster in the crown-of-thorns Acanthaster planci starfish. Baughman, Ken; Satoh, Nori; Shoguchi, Eiichi 73 A transcriptomic strategy for identifying novel mediators of the sea urchin larval immune response Buckley, Katherine M.; Ho, Eric; Rast, Jonathan 74 Expression of the karyopherin-alpha family of nuclear transport proteins in Lytechinus variegatus. Byrum, Christine; Smith, Jason; Easterling, Marietta; Bridges, M. Catherine 75 Transcriptome Analysis of Late Gastrula Pigment Cells Calestani, Cristina; Barsi, Julius C; Tu, Qian; Ortiz, Antonio; Buckley, Kate M; Stearnes, Ariel; Rast, Jonathan P; Davidson, Eric H 76 mTOR regulation of polysomal recruitment at fertilization Chassé, Héloïse; Boulben, Sandrine; Cormier, Patrick; Morales, Julia 77 SLC and Notch2 regulate dorsal-ventral PMC positioning and skeletal patterning Chung, Oliver; Piacentino, Michael; Hewitt, Finnegan; Patel, Vijeta; Ferrell, Patrick; Chaves, James; Li, Christy; Hameeduddin, Hajerah; Poutska, Albert; Bradham, Cynthia 78 Mesoderm and Micromere Development in Eucidaris tribuloides Coots, Ashley D.; Covington, Rae Ann; Lung, Kara; Wood, Maureen; Sweet, Hyla 79 Expression of embryonic skeletal development genes during adult arm regeneration of the brittle star Amphiura filiformis Czarkwiani, Anna; Dylus, David; Oliveri, Paola 80 Polarity of small micromeres and its impact on localization of plasma membrane proteins Espinoza, Jose A.; Campanale, Joseph; Gokirmak, Tufan; Hamdoun, Amro 81 Ophioplocus esmarki Embryo and Larvae Swimming Behavior Freyn, Alec W.; Coots, Ashley; Sweet, Hyla 82 Identifying and measuring voltage gradients in normal and perturbed embryos Hadyniak, Sarah E.; Schatzberg, Daphne; Lawton, Matthew L.; Bishop, Jacob; Beane, Wendy; Levin, Michael; Bradham, Cynthia 83 Semi-dry sea urchin experiment using preserved egg and sperm supply Kiyomoto, Masato 84 Development of the nervous system in a sea cucumber, Apostichopus japonicus: shift from bilateral to pentaradial symmetry Kondo, Mariko; Nagai, Akiko; Kikuchi, Mani; Omori, Akihito; Akasaka, Koji 85 Expression of germ cell markers in hemichordate Ptychodera flava: implication to the embryonic origin of PGCs in Ambulacraria Lin, Ching-Yi; Yu, Jr-Kai; Su, Yi-Hsien 86 BioTapestry: Interacting With the Network Directly in the Web Browser Longabaugh, William; Paquette, Suzanne; Leinonen, Kalle 87 How pluteus arms were evolved? Morino, Yoshiaki; Koga, Hiroyuki; Wada, Hiroshi Page 7 of 51 88 5-LOX is Required for Skeletal Patterning in Sea Urchin Embryos Murray, Ian S.; Patel, Vijeta; Li, Christy; Yu, Annie; Hameeduddin, Hajerah; Hewitt, Finnegan; Poustka, Albert; Bradham, Cynthia 89 Histological and molecular biological analysis on the adult nervous system of the feather star Oxycomanthus japonicus Omori, Akihito; Kurokawa, Daisuke; Akasaka, Koji 90 Circadian clock in the S. purpuratus larva: a diverged time-keeping mechanism that drives 24h rhythmicity? Petrone, Libero; Lerner, Avigdor; Oliveri, Paola 91 Appearance of Order Level Traits in the Triassic Echinoid Fossil Record Petsios, Elizabeth; Thompson, Jeffery; Bottjer, David 92 Concanamycin A perturbs dorsal-ventral specification in sea urchin embryos Reidy, Patrick; Bishop, Jacob; Schatzberg, Daphne; Zushin, Peter; Ross, Erik; Carney, Tamara; Bradham, Cynthia 93 LvBMP5-8 is required for normal skeletal patterning but not dorsal-ventral specification in the sea urchin embryo Ramachandran, Janani; Chung, Oliver; Piacentino, Michael; Reyna, Arlene; Yu, Jia; Hameeduddin, Hajerah; Poustka, Albert; Bradham, Cynthia 94 H+/K+ antiport activity is required for PMC differentiation and skeletogenesis Schatzberg, Daphne; Lawton, Matthew; Hadyniak, Sarah; Bishop, Jacob; Ross, Erik; Carney, Tamara; Beane, Wendy; Levin, Michael; Bradham, Cynthia 95 Genome-wide analysis of the skeletogenic gene regulatory network of sea urchins Shashikant, Tanvi; Rafiq, Kiran; Ettensohn, Charles 96 Expression of the ATP-Binding Cassette transporter Sp-ABCC5a in pigment cells is required for sea urchin gastrulation Shipp, Lauren E.; Hill, Rose; Moy, Gary; Gokirmak, Tufan; Hamdoun, Amro 97 microRNA-31 Regulates Skeletogenesis of the Sea Urchin Embryo Stepicheva, Nadezda; Song, Jia 98 miR-124 Regulation of the Delta/ Notch signaling pathway Suarez, Santiago N.; Song, Jia 99 Spatial Regulation of Gene Expression in the Skeletogenic Mesenchyme by Extrinsic Cues Sun, Zhongling; Ettensohn, Charles 100 A probabilistic modeling of the cell lineage highlights interindividual variability in Paracentrotus lividus early development villoutreix, paul; Rizzi, Barbara; Delile, Julien; Duloquin, Louise; Faure, Emmanuel; Savy, Thierry; Bourgine, Paul; Peyriéras, Nadine 101 The embryonic transcriptome for Lytechinus variegatus Zuch, Daniel; Hogan, J.D.; Keenan, Jessica; Luo, Lingqi; Saji, Akhil; Sundermeyer, Mary Ann; Piacentino, Michael; Schatzberg, Daphne; Azzizi, Elham; Zhang, Shile; Heilbut, Adrien; Poustka, Albert; Bradham, Cynthia Page 8 of 51 Talk Abstracts 1 Use of regulatory complexity in gut organogenesis Peter, Isabelle (Cal-Tech); Davidson, Eric; Valencia, Jonathan So far no gene regulatory network has been solved for any animal organogenesis process, the reason being both the enormous regulatory complexity underlying the formation of most organs and the challenges of experimental network analysis. The advanced state of GRN analysis in sea urchin embryos thus provides a unique opportunity to approach such large scale GRNs. We are particularly interested in solving the genomic program for gut development, because of its importance to animal development and evolution. GRN analysis for a process as complex as the formation of a compartmentalized gut however requires strategies different from the analysis of smaller pre-gastrular GRNs. To assess the dimensions of the gut GRN, we have analyzed the spatial expression of a majority of regulatory genes encoded in the sea urchin genome and expressed during the time from beginning of gastrulation up to 72h. The results show that more than 190 regulatory genes are expressed in the developing gut. The function of this so far unknown GRN is to specify within the anterior and posterior endoderm at least 25 distinct regulatory domains which pattern the mature gut at 72h. Thus even to form the relatively simple larval gut in sea urchin embryos requires a large part of the regulatory system encoded in the genome. 2 Spatial regulatory states in the ectoderm. McClay, David R. (Duke University) Two new territories of transcription factor expression have been identified in the ectoderm of the sea urchin embryo Lytechinus variegatus. The first region is a band of cells about 4 cells wide immediately adjacent to the endoderm. Wnt5 from the endoderm induces the border ectoderm via short range signaling. Nodal and BMP signaling then subdivides this band of ectoderm into dorsal and ventral zones. Perturbation experiments establish that the border ectoderm releases signals necessary for skeletal patterning. Especially important is a group of cells at the intersection of the border ectoderm and the dorsal-ventral margin. VEGF and FGF are produced at that intersection causing the ventrolateral cluster of PMCs to form immediately beneath, and causing initiation of skeletogenesis. The second territory of specialized gene expression is initiated by expression of ectodermal SoxC in a group of cells immediately above the ventral border ectoderm. That group of cells migrates to the ciliary band and differentiates as neurons. Perturbation of SoxC eliminates all neurons selectively, including the neurons originating in the ventral ectoderm. The ventral ectodermal SoxC pre-neurons also express achaete-scute and later express a series of known neural markers. This territory of pre-neural cell origin is distinct from the apical ectoderm where other neurons are known to originate. A large number of perturbation studies, guided in part by the ectodermal gene regulatory network model, establish SoxC as an early transcription factor in pre-neural cells. 3 A maternal determinant of the dorsal-ventral axis in Paracentrotus lividus? Lepage, Thierry (CNRS); Haillot, Emmanuel (Institute of Biology Valrose) Classical studies showed that, unlike the animal-vegetal axis, the dorsal-ventral axis of the sea urchin embryo is not pre-established rigidly in the egg. Consistent with this idea, specification of the dorsal-ventral axis critically relies on cell interactions and on the zygotic expression of nodal. Nodal expression starts around the 32-60-cell stage and is first very broad, before being restricted to the Page 9 of 51 ventral ectoderm. It has been proposed that Redox gradients generated by asymmetrically distributed mitochondria, and possibly acting through p38, may provide the initial spatial cue that initiates nodal expression or alternatively, that Redox gradients may be required for the spatial restriction of nodal. However, manipulating redox gradients has only modest effects on the orientation of the D/V axis and how the polarized expression of nodal is established is not well understood. We discovered a maternal factor that we named Panda (Paracentrotus Anti Nodal Dorsalizing Activity) that directs the orientation of the D/V axis by controlling the spatial expression of nodal. panda transcripts are broadly distributed, with a slight gradient present in a fraction of the embryos. Embryos injected with morpholino oligonucleotides targeting maternal panda transcripts failed to restrict nodal expression. Misexpression of panda oriented the dorsal-ventral axis in nearly 100% of the injected embryos. Finally, rescue experiments demonstrated that the activity of Panda is required locally in the early embryo. These findings identify Panda as a maternal factor that is both required for the spatial restriction of nodal and sufficient to orient the dorsal-ventral axis when misexpressed. Panda therefore fulfils some of the requirements for a maternal determinant of the D/V axis. These results suggest that, although the dorsal-ventral axis of the sea urchin embryo is not rigidly fixed, this axis may be prefigured in the egg in the form of a broad gradient of this maternally deposited factor. 4 Sea urchin GRN model Davidson, Eric (CalTech) The sea urchin embryo GRN up to gastrulation has now been expanded to include all but the apical neurogenic domain and the mesenchyme blastula stage oral/aboral mesoderm domains. Thus it becomes possible to consider the form of a global, genomically encoded network that encompasses the majority of a developing organism. I briefly discuss both the recurrent overall characteristics of the global developmental program explicit in this GRN, and the interesting special features of certain of its subregions. 5 Developmental systems drift: making similar embryos with divergent genomes. Lemaire, Patrick (CRBM, UMR5237, CNRS/University Montpellier; Institut de Biologie Computationnelle, Montpellier, France) The relationships between genotype and phenotype during evolution are complex and poorly understood. Surprisingly dissimilar genotypes and developmental programmes can translate into very similar phenotypes. This may explain how some species can remain morphologically similar for long periods of time in spite of extensive genome divergence. It also highlights that morphological similarity does not necessarily reflects molecular homology, a phenomenon that should be taken into consideration when extrapolating to Man results obtained with mammalian model organisms. Ascidian embryos constitute a remarkable system to study morphological stasis. Slow evolution of their stereotyped embryonic morphologies, based on invariant cell lineages, allows comparison of the same developmental processes across hundreds of millions of years. Extreme genome intra-specific polymorphism and inter-specific divergence suggests an astounding level of plasticity in the underlying developmental pathways. During the talk, I will present the computational tools that we are developing in collaboration with the Virtual PLant research team (Montpellier) to quantify embryonic morphologies and their variability from light-sheet microcopy recordings of live embryos. I will also give a preliminary assessment of genome and transcriptome divergence within and between ascidian genera. I will conclude by presenting cis-regulatory mechanisms that explain, in part, how divergent genomes can support morphological invariance. Page 10 of 51 6 Embryonic Neurogenesis Burke, Robert D. (University of Victoria) Urchins appear to share many of the common features of neurogenesis of metazoan embryos. The morphological aspects of the differentiation phase of neurogenesis in urchin embryos are documented, however, our understanding of neural specification and the regulation of proneural networks is fragmentary. This is in part due to a lack of early markers for neural progenitors. Sox B2 is expressed broadly in oral ectoderm and ciliary band and some, but not all Sox B2 expressing cells become neurons. Six3 is necessary for neural specification, but expression is transitory and it does not serve to identify neural progenitors. In blastulae, Sox C is expressed in vegetal mesendoderm cells of the animal pole domain, and during gastrulation it appears in cells scattered throughout the oral ectoderm. SoxC has several of the features of a marker for neural progenitors. Delta-Notch signaling is a common feature of metazoan neurogenesis that produces committed progenitors and it appears to be a critical phase of neurogenesis in urchin embryos. We are attempting to determine the critical functions of these neurogenic proteins to develop a model that articulates the gene regulatory networks with the cellular context in which neural development occurs. Urchins are a facile, but powerful model with the potential of revealing many shared and derived features of deuterostome neurogenesis. 7 Gradients and networks that pattern the Drosophila embryo Small, Stephen (New York University); Chen, Hongtao (New York University); Ochoa-Espinosa, Amanda (New York University); Xu, Zhe (New York University); Cao, Jinshuai (New York University) The early Drosophila embryo develops as a syncytium of nuclei, and spatial patterning of the embryo is controlled by a well-characterized network of transcription factors. Here we focus on the patterning activities of the Bicoid transcription factor, which is distributed in a long-range gradient along the anterior posterior axis. Previous studies suggest that Bcd functions as a “morphogen”, which establishes multiple gene expression boundaries by concentration-dependent activation mechanisms. We have used in vivo gradient manipulations and an extensive analysis of Bcd-dependent enhancers to critically test this hypothesis. Our results strongly suggest that most target genes do not respond to specific concentration thresholds within the gradient. Rather, target gene boundaries are positioned primarily by repressive gradients that antagonize Bcd-dependent activation. Thus, embryo organization is more accurately described as an emergent property of the whole network of interacting genes. 8 Pathways Regulating Stem Cell Induction, Self-Renewal and Engraftment Zon, Leonard (HHMI/Boston Children's Hospital) Hematopoietic stem cell transplantation involves the homing of stem cells to the marrow, an active process of engraftment, and the self-renewal of the blood stem cells. We have been using the zebrafish as a model to study the molecular biology of this process. By imaging RUNX1 GFP+ cells arriving in the next site of hematopoiesis (the caudal hematopoietic territory), engraftment can be visualized. This process involves an attachment phase and then an extravasation to the abluminal side of the endothelial cells. The endothelial cells cuddle the hematopoietic stem cell and the stem cells have the ability to be maintained in a quiescent fashion or to divide symmetrically or asymmetrically. Using chemical screens, we have found small molecules that can enhance engraftment or suppress engraftment. We also have developed a new technique of culturing zebrafish blastomeres and examining tissue differentiation. Our initial work led to a chemical screen that demonstrated that activation of the FGF, wnt, and cAMP pathways stimulate muscle development. A cocktail of chemicals found in our zebrafish culture assays was sufficient to reprogram human iPS cells to skeletal muscle. The system has now been used to reprogram neural crest cells and blood stem cells. Our studies have found fundamental aspects of stem cell biology that may be therapeutically useful for patients. Page 11 of 51 9 Chuck Ettensohn 10 Athula Wikramanayaka 11 Amro Hamdoun 12 Evolution of post transcriptional mechanisms for germ line segregation in echinoderms Swartz, S. Zachary (Brown University Molec Biology, Cell Biology & Biochemistry); Reich, Adrian (Brown University); Oulhen, Nathalie (Brown University); Raz, Tal (Helicos Biosciences); Milos, Patrice (Helicos Biosciences); Campanale, Joseph (UCSD Scripps); Hamdoun, Amro (UCSD Scripps); Wessel, Gary (Brown University) A critical event in animal development is the specification of primordial germ cells (PGCs), which become the stem cells that create sperm and eggs. Germ line segregation can be categorized within a continuum of inherited and inductive mechanisms. The inherited mode, typified by organisms such as the fruit fly Drosophila, involves spatial localization of maternally supplied molecular determinants, collectively called a germ plasm. The embryonic cells that inherit this material are thus directed toward germ line fate. Animals such as the mouse do not employ a germ plasm, but rather rely upon inductive signaling between tissue layers to specify germ cells. We find that the PGCs of the sea urchin Strongylocentrotus purpuratus exhibit broad transcriptional repression, yet enrichment for a set of inherited mRNAs. Enrichment of several germ line determinants in the PGCs requires the RNA binding protein Nanos to deplete the transcript encoding CNOT6, a deadenylase, in the PGCs, thereby creating a stable environment for RNA. Alteration of CNOT6 levels in the PGCs and somatic cells results in their failure to selectively retain Seawi transcripts and Vasa protein. We propose a "time-capsule" model of germ line determination and continuity of a cryptic germ plasm in the sea urchin, which has classically been thought of as an inductive embryo. Comparisons with the pencil urchin, sea star, and sea cucumber suggest Nanos depletion of CNOT6 is a conserved mechanism within echinoderms. In addition to this primary inherited mechanism, the sea urchin embryo is capable of compensating for the loss of its germ line under certain conditions. We identify z426, a maternally supplied putative RNA binding protein, as a potential mediator for this process. 13 Quantification of Exocytosis Kinetics by DIC Image Analysis of Cortical Lawns Poccia, Dominic L. (Amherst College) Cortical lawns prepared from sea urchin eggs (Vacquier in Dev Biol 43:62–74, 1975) have served for many years as a robust in vitro system for study of regulated exocytosis and membrane fusion events. Lawns have been imaged by various microscopy techniques and quantification of exocytosis kinetics has been achieved primarily with specialized light scattering techniques and flow cells. We present simple differential interference contrast image analysis procedures for quantifying the kinetics and extent of exocytosis in cortical lawns using an open vessel that allows rapid solvent equilibration and modification. These preparations maintain biologically relevant architecture of the original cortices, allow for cytological and immunocytochemical analyses, and permit quantification of variation within and between lawns. This simple technique should allow investigations into the contributions of proteins and phospholipids of endoplasmic reticulum, granule and plasma membranes in regulation of exocytosis. 14 Hedgehog signaling functions during sea star development Zazueta-Novoa, Vanesa (Brown University); Wessel, Gary (Brown University) The Hedgehog pathway controls a wide variety of developmental processes. One such role for Hh signaling is in the development of the gut during embryonic development. Hedgehog (Hh) is a secreted Page 12 of 51 protein that is enzymatically modified to make it active and it binds to its receptor, Patched (Ptch). In absence of Hh, Ptch acts as an inhibitor, binding and blocking Smoothened (Smo) to activate the pathway. When Hh is present, it binds to Ptch, releasing the inhibition of Smo. Smo antagonizes kinesin-related protein Costal 2 (Co2) activity, preventing the transcription factor cubitus interruptus (Ci in Drosophila or Gli in vertebrates) from cleavage. Then, full-length Ci protein proceeds to the nucleus where it activates transcription. In sea urchin, Hh expression profiles showed that Hh transcripts are enriched in the endoderm, suggesting that Hedgehog signal is produced by the endodermal tissue of the archenteron and function in formation of the mesodermal tissues. By addition of cyclopamine, sea urchin embryos revealed several defects, but the most relevant was the disruption of coelomic pouch morphogenesis. In cyclopamine-treated embryos, coelomic pouch cell progenitors at the tip of the archenteron did not split in two groups and relocate laterally to form the coelomic pouches as they do in normal conditions. Instead these cells remained atop the archenteron and formed one pouch-like structure. We studied the Hh signaling pathway in sea star and we identified two Hh sequences, two Ptch sequences and one Smo sequence using a de novo transcriptome database. We analyzed their expression profiles and the effect on early development using cyclopamine. At lower concentrations, cyclopamine incubation of the embryos caused a general delay in the development of the larva and at higher concentrations, it caused defects in gut formation or death of the larvae as it was reported previously in sea urchin. However, defects in archenteron and gut patterning are being addressed. 15 Nanos protein is specifically retained in the small micromeres. Oulhen, Nathalie (Brown University); Wessel, Gary (Brown University) Nanos is a translational regulator required for the survival and maintenance of primordial germ cells during embryogenesis. It is usually expressed uniquely in germ cells, and is often "toxic" if active elsewhere in the embryo. Three nanos homologs are present in the genome of the sea urchin Strongylocentrotus purpuratus (Sp),each nanos mRNA accumulates specifically in the small micromeres (SMM), the lineage that contributes to the germline. Sp nanos2 3’UTR is sufficient for RNA retention and protein accumulation selectively in the SMM lineage. We found that an additional mechanism regulates Sp nanos2 expression. Sp nanos2 ORF also leads to a selective reporter enrichment in the small micromeres, independently of the 3’UTR. Our results suggest that post translational modifications such as sumoylation, ubiquitination, or acetylation do not affect nanos expression. Moreover, mutations in nanos zinc finger domains do not affect the protein stability in the SMM. We previously showed that nanos expression is controlled by a combination of selective RNA retention and translational control mechanisms, we are now adding an additional level of regulation affecting its protein stability. 16 Sea urchins as models for the construction of prototypic embryonic development Peyrieras, Nadine (CNRS); Fabreges, Dimitri (CNRS); Barbara, Rizzi (CNRS); Villoutreix, Paul (CNRS); Delile, Julien (CNRS); Savy, Thierry (CNRS); Duloquin, Louise (CNRS); Suret, Pierre (Lille University); Doursat, René (Drexel Unviversity); Bourgine, Paul (CNRS) On the way to a better understanding of embryonic development, we aim at systematizing a quantitative and modeling approach of developing organisms based on live imaging and automated image processing. Sea urchins should be ideal models for a proof of concept, thanks to their egg accessibility, small size, rapid embryonic development and relative transparency. Most importantly, several species have been extensively explored at the cellular, genetic and molecular level, and an impressive amount of data is available to be assimilated in integrative models. Immobilization of the embryos to image them in toto by state of the art 2-photon laser scanning or selective plane imaging microscopy, remains however a major issue. Our current model explored the space of phenotypic features in Paracentrotus Page 13 of 51 lividus at blastula stages. A small cohort of individuals was used to propose probabilistic models for cell behavior and their integration in a spatial multi agent model was developed to assess biomechanical constraints shaping the blastula. The comparative study of Paracentrotus lividus and Sphaerechinus granularis aims at quantifying intra- and inter-specific variability. S. granularis has the advantage to undergo primary mesenchymal cells ingression prior swimming. It thus appears as an interesting model for the quantitative approach of the variation components in the twin paradigm. The development of twins obtained by the classical separation of 2-cell stage blastomeres challenges our understanding of normal development. Again, the need for cell lineage analysis points to the usefulness of a quantitative automated approach based on live imaging. We expect that the community will synergize in exploiting an open source database of digital specimens and make it grow by using the BioEmergences webservice for 3D+time data processing. 17 ATP-binding cassette transporters mediate small micromere migration and left/right coelomic pouch segregation in Strongylocentrotus pupuratus Campanale, Joseph P. (Scripps Institution of Oceanography); Espinoza, Jose (Scripps Institution of Oceanography); Gokirmak, Tufan (Scripps Institution of Oceanography); Hamdoun, Amro (Scripps Institution of Oceanography) One function of ATP-binding cassette (ABC) transporters is to establish morphogenetic gradients in embryos. The small micromeres (Smics) undergo a 65% reduction in ABC-transporter activity at formation, presumably to concentrate morphogenetic signals necessary for segregation between the two coelomic pouches (CPs). In controls, an average of five Smics are found in the left coelomic pouch (CP), while embryos grown in the presence of ABC-transporter inhibitors have random left/right Smic distributions. To test how ABC transporters control segregation, we fused Sp-ABCB1a to Sp-nanos UTRs and selectively overexpressed it in Smics. In these embryos a dose-dependent increase in the number of Smics in the left CP was observed, while targeted expression of membrane markers and null mutants of ABCB1a did not alter Smic distribution. These results suggest that the expression of ABCB1a in Smics disrupts their left/right migration, possibly by dampening the detection of chemoattractants. We recently showed that Smics transition from non-motile epithelial cells to motile quasi-mesenchymal cells during gastrulation. Smics in gastrulae are embedded in the tip of the archenteron, but remain motile and develop cortical blebs and dynamic filopodia that contact ectoderm. Smics in prism larvae move in the plane of the blastoderm towards the coelomic pouches. Using this information, we are currently investigating whether ABC-transporter activity disrupts Smic migration by altering bleb and filopodia dynamics. Preliminary results indicate Smics expressing ABCB1a in vitro extend smaller blebs and fewer filopodia than Smics expressing non-transporter membrane proteins. Collectively, our findings suggest that ABC-transporter mediated secretion of morphogens controls Smic migration leading to their left/right patterning. We speculate that Smic left/right patterning is linked to ABC-transporter mediated Smic migration dynamics. 18 Inhibition of the Arp2/3 Complex in Sea Urchin Coelomocytes Induces a Lamellipodial to Filopodial Shape Change and Alters the Cell Spreading Process Henson, John (Dickinson College); Goldson, Brandon (Dickinson College); Patterson, Rebecca (Dickinson College); Shen, Eileen (Dickinson College); Brown, Briana (Dickinson College); Medrano, Angela (Dickinson College) Our previous studies on sea urchin coelomocytes have indicated that Arp2/3 complex-facilitated actin filament polymerization is crucial for structuring the dendritic actin filament network characteristic of the peripheral lamellipodial region, for generating the pushing force component of actin-based centripetal flow, and for closing cytoplasmic wounds. In the present study live cell, fluorescence, and 3D structured Page 14 of 51 illumination super-resolution light microscopy, as well as critical-point-dry and rotary shadow transmission electron microscopy were used to study the impact of treating coelomocytes in suspension with the specific inhibitor of the Arp2/3 complex, CK666. In adherent cells treated with CK666 the dendritic network of actin filaments present in the lamellipodium is replaced by an array of transversely oriented arcs of elongate actin filaments. In contrast, when coelomocytes are treated in suspension with CK666 they undergo a radical morphological transformation from a lamellipodial to highly filopodial form. In addition, when these filopodial cells are allowed to settle onto a glass substrate their spreading process proceeds via a novel structural reorganization of actin involving the presence of concentric circle patterns of elongate actin filaments in regions of filopodial spreading. Comparison of these cells with control cells in which spreading takes place in the absence of CK666 indicates that the rate of the overall spreading process is also significantly slowed by the presence of the inhibitor. These results demonstrate that Arp2/3 complex-facilitated actin polymerization is essential for the maintenance of the lamellipodial form of coelomocytes in suspension and for the normal spreading of these cells on a substrate. They also indicate that cells retain the ability to spread even in the absence of the machinery necessary to generate the dendritic array of branched actin filaments present in lamellipodia. 19 General approach for in vivo recovery of cell type specific effector gene sets Barsi, Julius C. (Caltech); Tu, Qiang (Caltech); Davidson, Eric (Caltech) Differentially expressed, cell type specific effector gene sets hold the key to multiple important problems in biology, from theoretical aspects of developmental gene regulatory networks (GRNs) to various practical applications. Although individual cell types of interest have been recovered by various methods and analyzed, systematic recovery of multiple cell type specific gene sets from whole developing organisms has remained problematical. My presentation will delineate general methodology aimed at obtaining this biological information and is applicable to all model organisms for which transgenic tools are available. It has been devised using the sea urchin embryo, material of choice because of the large-scale GRNs already solved for this model system. The method utilizes the regulatory states expressed by given cells of the embryo to define cell type, and includes a Fluorescence Activated Cell Sorting (FACS) procedure that results in no perturbation of transcript representation. I have extensively validated the method by spatial and qualitative analyses of the transcriptome expressed in isolated embryonic skeletogenic cells and as a consequence, generated a prototypical cell type specific transcriptome database: http://www.spbase.org:3838/cellspecific/ 20 An anterior signaling center patterns and sizes the anterior neuroectoderm territory of the sea urchin embryo Range, Ryan (Mississippi State University); Angerer, Robert (NIDCR, National Institutes of Health); Angerer, Lynne (NIDCR, National Institutes of Health) Anterior signaling centers are essential to specify and pattern the early anterior neuroectoderm (ANE) in several deuterostome embryos. In the sea urchin embryo, the early ANE is restricted to the anterior end of the late blastula-stage embryo where it separates into inner and outer regulatory domains expressing the cardinal ANE transcriptional regulators, FoxQ2 and Six3, respectively. This patterning process is driven by FoxQ2, which is required to eliminate expression of six3 from the inner domain. FoxQ2 also activates the expression of two secreted Wnt regulators, sFrp1/5 and Dkk3, the activities of which define the correct sizes of the inner and outer ANE territories. Furthermore, the levels of sFrp1/5 and Dkk3 are rigidly maintained via auto-repressive and cross-repressive interactions with Wnt signaling components and ANE transcription factors. Our data support a model in which Six3 and FoxQ2 establish an anterior patterning center that ensures correct ANE patterning and border positions. Comparisons of functional Page 15 of 51 and expression studies in sea urchins, hemichordates and vertebrates show striking similarities in deuterostome ANE regulatory states and the molecular mechanisms that position and define its borders. These data provide strong support for the idea that the sea urchin embryo uses an ancient anterior patterning system that was present in the common ambulacrarian/chordate ancestor. 21 A Comprehensive Survey of wnt and frizzled Expression in the Sea Urchin Paracentrotus lividus Croce, Jenifer (CNRS); Robert, Nicolas (UPMC); Lhomond, Guy (CNRS); Schubert, Michael (CNRS) WNT signaling is, in all multicellular animals, an essential intercellular communication pathway that is critical for shaping the embryo. At the molecular level, WNT signals can be transmitted by several transduction cascades, all activated commonly by the binding of WNT ligands to receptors of the FRIZZLED family. The first step in assessing the biological functions of WNT signaling during embryogenesis is thus the establishment of the spatiotemporal expression profiles of wnt and frizzled genes during development. To this end, using qPCR, Northern blot and in situ hybridization assays, we have thus determined the comprehensive expression patterns of all eleven wnt and four frizzled genes present in the genome of the sea urchin Paracentrotus lividus throughout its embryogenesis. Our findings indicate that the expression of these wnt ligands and frizzled receptors is highly dynamic in both time and space. We further establish that all wnt genes are chiefly transcribed in the vegetal hemisphere of the embryo, whereas expression of the frizzled genes is distributed more widely across the embryonic territories. Thus, in P. lividus, WNT ligands might act both as short- and long-range signaling molecules that may operate in all cell lineages and tissues to control various developmental processes throughout embryogenesis. 22 Pigment cell migration: The role of Eph-Ephrin signaling Krupke, Oliver (University of Victoria) Studies on mesenchyme specification in the sea urchin have produced one of the most clearly defined gene regulatory networks to date and there is an ongoing effort to characterise effectors that relay this genetic information to produce morphological changes and cell movements that further define cell fate. Following specification and epithelial-mesenchyme transition, mesenchyme cells ingress into the embryo and migrate to specific embryonic regions where they differentiate to skeletonogenic or non-skeletogenic fates. Pigment cells are specialized, echinochrome-containing, non-skeletogenic secondary mesenchyme with apparent roles in innate immunity. Using targeted gene knockdown, direct inhibition of Eph kinase and ectopic expression of Ephrin, we show Eph-Ephrin signaling is necessary and sufficient for pigment cell migration, directing them from within the blastocoel to positions adjacent the aboral basement membrane where they extend processes into the overlying ectoderm. Using immunofluorescence, we identify a putative mesenchyme-epithelial transition in pigment cells following contact with the basal lamina and pigment cells exhibit a number of features characteristic of an MET event. Combined our results highlight a developing model for MET wherein the upstream and downstream components can be visualized in an easily manipulated embryo. 23 Mechanisms of Small Micromere Homing Martik, Megan L. (Duke University); McClay, David R. (Duke University) The small micromeres arise at the vegetal pole from an unequal 5 th cleavage, and their progeny are specified to become the primordial germ cells. Throughout gastrulation, small micromeres actively extend filopodia and lamellipodia at the tip of the gut though they continue to remain part of the invaginating epithelium and express LvG-cadherin. Once gastrulation nears completion, the tip of the gut Page 16 of 51 undergoes basement membrane remodeling that facilitates the small micromeres’ epithelial-mesenchymal transition (EMT) and migration over the archenteron to the posterior halves of the forming coelomic pouches. We show that the small micromeres reach the coelomic pouches via a directed homing mechanism. Ectopically placed small micromeres are able to find their way home to the coelomic pouches from any starting position in the embryo. Spatially and temporally misplaced 16-cell stage micromeres also home upon insertion into the blastocoel of a late gastrula host. The small micromeres home directly to the non-skeletogenic mesoderm (NSM) lineage of the forming coelomic pouch. Small micromeres are directed to the NSM via evolutionarily conserved signaling and chemoattractant mechanisms. When the NSM is not specified, the small micromeres are unable to home. Transcription factor knockdowns within the NSM lineage shed light on the regulatory underpinnings of homing mechanisms. By using the robust homing behavior of these cells, we uncovered a transcriptional circuitry responsible, in part, for directed homing mechanisms of germ cells. Current aims are to connect the underlying transcriptional regulation of the signaling with the chemoattractant mechanisms by which primordial germ cells undergo such a dramatic feat of finding their way home. 24 Transcriptional control of immune cell development in the purple sea urchin embryo Schrankel, Catherine S. (University of Toronto) Immune response in the purple sea urchin larva is mediated by granular pigment cells and several derivatives of the blastocoelar cells. Blastocoelar immunocytes develop from an oral patch of NSM cells in the 24-hr embryo that co-express orthologs of the vertebrate hematopoietic stem cell transcription factors Gata1/2/3, Scl/Tal-2/Lyl-1 and Erg/Fli-1. Upon down-regulation of these genes, immunocyte precursors initiate epithelial-to-mesenchymal transition (EMT), undergo limited cell division and begin to express subsets of immune differentiation markers. When SpGata123 is perturbed, precursors fail to migrate and terminal gene expression is greatly reduced. SpGata123 itself appears to provide negative feedback into this system (as does Gata1 in vertebrate stem cells). Thus in the sea urchin embryo, the co-expression of SpGata123, Scl, Erg/Fli-1 may affect a transient multipotent state that switches towards immune differentiation upon the down-regulation. Targets of SpGata123 in immunoctye precursors may include EMT genes and upstream regulators of terminal gene batteries. Class I bHLH transcription factors also play a role in this system, including SpE-protein, the single homolog of vertebrate E2A/HEB/ITF2 that is the relevant binding partner of Scl, and SpId, a negative regulator of class I BHLH activity. We have identified an alternative start in SpE-protein, also found in vertebrate HEB and ITF2. This is the first such domain to be characterized in an invertebrate. SpEalt is expressed specifically and dynamically in subsets of the developing NSM. As sea urchins are evolutionarily situated at the base of the deuterostomes, the regulatory links between SpGata123 down-regulation and E-protein activity to programs of immunocyte EMT and differentiation may be conserved in a minimal gene network that is ancient to deuterostome immune cell development. 25 Insights from amphioxus and lamprey into the evolution of vertebrate head skeleton development Medeiros, Daniel (University of Colorado, Boulder) A defining feature of vertebrates (craniates) is a pronounced head supported and protected by a robust, cellular endoskeleton. In the first vertebrates, this skeleton was likely built of collagenous cellular cartilage, which forms the embryonic skeleton of all vertebrates and the adult skeleton of modern jawless and cartilaginous fish. In the head, most cellular cartilage is derived from the neural crest, a migratory cell population that arises from the edges of the central nervous system (CNS). Because collagenous Page 17 of 51 cellular cartilage and neural crest cells (NCC) have never been described in invertebrates, the appearance of NCC-derived cellular cartilage is considered a turning point in vertebrate evolution. Using new methods for the continuous lab culture of the cephalochordate Branchiostoma floridae, we find that a skeletal tissue displaying essentially all of the core conserved features of vertebrate cellular cartilage forms transiently during metamorphosis of this species. We also present evidence that a key regulator of collagenous cellular cartilage development, SoxE, gained new cis-regulatory sequences during vertebrate evolution to direct its novel expression in NCC. Together these results suggest that the origin of the vertebrate head skeleton was not dependent on the evolution of a qualitatively new skeletal tissue, but on the spread of this tissue throughout the head. We further propose that the evolution of cis-regulatory elements near an ancient regulator of cartilage differentiation was a major factor in the evolution of the vertebrate head skeleton. 26 Ophiuroid skeletogenesis in development and regeneration Oliveri, Paola (University College London) Calcitic endoskeleton is a fundamental character of echinoderms. In all adults it is represented as mesh-like ossicle units, while in larvae it is prominently formed as radiate spicules only in two classes: Echinoidea and Ophiuroidea. The regulatory program for the development of the sea urchin larval skeleton has been extensively studied and provides an excellent framework to study evolution of regulatory networks governing cell type specification. Very little is known about skeletogenesis in ophiuroids. I will describe the effort we are undertaking to develop molecular resources and experimental approaches to study skeletogenesis during the development and adult arm regeneration of the ophiuroid Amphiura filiformis. From a developmental transcriptome, we isolated and studied the spatio-temporal expression of more than 25 genes orthologous of sea urchin developmenatal skletogenic (SM) and non skeletogenic mesodermal (NSM) genes. By early blastula stage, a group of cells, within the vegetal plate, show a SM molecular signature by expressing genes such as alx1 and p19. Later they will be the first ingressing cells and will secrete two bilaterally arrayed spicules. Despite a rather similar embryonic development compared to sea urchin, A. filiformis shows several differences in regulatory states of both SM and NSM cells, suggesting different network linkages. During arm regeneration different skeletal elements are formed at different moment of the regeneration process. Cells in sub-epithelial position in the blastema, where skeleton primordial are formed, show similar regulatory states to the embryonic SM cells, indicating the usage of a similar embryonic SM network module. On the contrary, at later stages of regeneration different skeletal elements have different regulatory states. 27 The role of Nodal signaling in axis formation of the hemichordate Saccoglossus kowalevskii Lowe, Chris (Stanford University); Wlizla, Marcin (University of Chicago); Gonzalez, Paul (Stanford University); Darras, Sebastien (Observatoire Oceanologique de Banyuls) The role of Nodal signaling during the early embryonic development of chordates and echinoderms is well characterized. This ligand plays key roles in the early establishment of the embryonic axes in both groups. While there are similarities in the role of Nodal signaling in dorsoventral and left/right patterning between groups, further comparative data from hemichordates are required for a more comprehensive understanding of the evolution of the this signaling pathway during early deuterostome evolution. We present functional data on the role of Nodal signaling in the direct-developing hemichordate Saccoglossus kowalevskii during early axis formation. There are three Nodal ligands in S. kowalevskii representing a recent diversification of this ligand family. Two of the ligands are expressed in the endomesodermal precursors at early blastula, and a third in the prospective posterior ectoderm in later Page 18 of 51 blastula and through gastrulation. We present both specific knockdown by RNAi and inhibitor experiments, and over-expression by mRNA injection and protein incubations, to disrupt Nodal signaling and assay effects on early axis formation. 28 Cis-reg logic of Ese and Prox genes underlying output in blastocoelar mesoderm founders. Ransick, Andrew (Caltech); Davidson, Eric (Caltech) In euechinoids, the induction mechanism which initially specifies the chromogenic mesoderm founders occurs relatively early compared to establishment of the other indispensible non-skeletogenic mesodermal cell types of the pluteus larva, which include the pleisiomorphic mesodermal lineages for muscle cells, blastocoelar mesenchyme and coeloms. Two significant consequences of this specification sequence for the architecture of the mesodermal GRN are, firstly, that primary non-skeletogenic mesodermal territory founders all activate the regulators GCM and GataE, which initially biases them toward the pigment cell fate. Second, mechanism(s) must exist to redirect specification of a portion of these non-skeletogenic mesodermal founders toward the other non-pigment cell fates mentioned above. Cis-regulatory analyses of the regulators Ese and Prox, which are expressed exclusively in this secondarily established cohort of non-pigment cell mesodermal founders, are providing insights into the early mesoderm GRN that directs establishment of mesodermal sub-lineages. 29 Asymmetrical stabilization of hypoxia inducible factor a during sea urchin embryogenesis Su, Yi-Hsien (Institute of Cellular and Organismic Biology, Academia Sinica) Establishment of the dorsal-ventral axis during early development is a crucial step for bilaterians to build their bilateral symmetric bodies. In sea urchin embryos, unequal distribution of mitochondria is important for patterning their oral/ventral-aboral/dorsal axis. The prospective oral side contains more mitochondria and is more oxidizing than the aboral side. Hypoxia has been shown to impair the specification of the axis and result in a radially symmetric embryo. Hypoxia inducible factor α (Hifα) is a transcription factor that is stabilized when cells are exposed to hypoxia whereas it is degraded under normaxia when its proline residues are hydroxylated by hydroxylase enzymes. Previously we demonstrated that sea urchin Hifα (SpHifα) initiates the gene regulatory network controlling the specification of aboral ectoderm. However, SpHifα transcript is maternally deposited and distributed ubiquitously in the early embryo and how its gene product selectively activates the aboral genes is unknown. Here we show that SpHifα is asymmetrically stabilized and activated on the aboral side of the blastula embryo. Hypoxia and hydroxylase inhibitor DMOG both abolish the asymmetrical distribution of SpHifα. SpHifα proteins also become symmetrically distributed when their proline residues are mutated. Within embryo clusters that the redox gradient is re-established from the inside to the outside, SpHifα proteins are predominantly localized in the inside that later tends to become the aboral side of the embryo. These results suggest that the hydroxylation and degradation machinery acting on SpHifα is activated on the oral side of the embryo and the asymmetrical stabilization of SpHifα on the aboral side is regulated by the redox gradient. The differential activation of Hifα in the sea urchin embryo thus provides an example of its fundamental role during normal embryogenesis. 30 Gene Regulatory Network Governing 2-Dimensional Expression Patterns in the Sea Urchin Ectoderm Li, Enhu (Caltech); Cui, Miao (California Institute of Technology); Peter, Isabelle (California Institute of Technology); Davidson, Eric (California Institute of Technology) Regulatory state boundary formation is a general process in early development, in which embryonic territory is divided up into spatial domains which express distinct sets of regulatory genes. We establish Page 19 of 51 the mechanistic principles by which multiple orthogonal boundaries of this kind are progressively formed on the oral side of the sea urchin embryo, according to an encoded genomic program. These boundaries separate prospective endoderm from ectoderm domains, neurogenic from non-neurogenic domains, ciliated band from oral ectoderm domains, and produce an orthogonal grid of regulatory states. Boundary formation invariably depends on spatial transcriptional repression superimposed on more widespread domains of transcriptional activation. 31 A global assessment of Wnt family functions in the sea urchin embryonic gene regulatory networks Cui, Miao i. (Caltech) Wnt signaling affects cell fate specification processes throughout development. Here we take advantage of the well-studied gene regulatory networks (GRNs) for sea urchin embryogenesis to investigate the regulatory functions of the Wnt signaling system during pre-gastrular development. We show that all five expressed wnt genes display similar features of expression: spatially dynamic early expression, later restriction to posterior endoderm and/or vegetal ectoderm and complete absence from the animal half of the embryo. Expression of frizzled genes is largely non-overlapping, in sum covering most domains of the embryo. Wnt signaling is not required during early development, due to maternal nuclear b-catenin, but later specifically regulates endodermal and apical neurogenic GRNs, as shown by monitoring genome-wide regulatory gene expression in embryos treated with the C59 Wnt signaling inhibitor. Morpholino perturbations of individual Wnts demonstrates their functional divergence, such that Wnt8 restricts apical neurogenic fates and Wnt1 and Wnt16 activates endodermal regulatory genes. A positive feedback circuit between wnt1, wnt16 and hox11/13b highlights the importance of Wnt signaling in the posterior endoderm. In addition, we show the specific regulatory functions of Wnt ligands in embryonic patterning along the primary vegetal-animal axis. This work thus reveals both comprehensive and particular functions of Wnt signaling in the global regulatory context of this embryo. 32 Carmen Andrikou 33 New Genes in Dorsal-Ventral Skeletal Patterning Bradham, Cynthia (Biology Department, Boston University); Piacentino, Michael (Biology Department, Boston University); Zuch, Daniel (Biology Department, Boston University); Hewitt, Finnegan (Biology Department, Boston University); Ramachandran, Janani (Biology Department, Boston University); Chung, Oliver (Biology Department, Boston University); Reyna, Arlene (Biology Department, Boston University); Hameeduddin, Hajerah (Biology Department, Boston University); Li, Christy (Biology Department, Boston University); Yu, Jia (Biology Department, Boston University); Patel, Vijeta (Biology Department, Boston University); Chaves, James (Biology Department, Boston University); Ferrell, Patrick (Biology Department, Boston University); Bardot, Evan (Biology Department, Boston University); Lee, David (Biology Department, Boston University); Shaw, Scott (Biology Department, Boston University); Cho, Ah Ra (Biology Department, Boston University); Core, Amanda (Biology Department, Boston University); Tse, Matt (Biology Department, Boston University); Olenik, Ekaterina (Scientific Computing and Visualization, Boston University); Keenan, Jessica (Program in Bioinformatics, Boston University); Hogan, J.D. (Program in Bioinformatics, Boston University); Luo, Lingqi (Program in Bioinformatics, Boston University); Coulomb-Huntington, Jasmin (Program in Bioinformatics, Boston University); Poutska, Albert J. (Max-Planck Institut fuer Molekulare Genetik, Berlin) Skeletal patterning in sea urchin embryos is regulated by cues within the ectoderm that direct the position of the skeleton-secreting PMCs. We performed an RNA seq-based screen to identify those cues based on the hypothesis that patterning genes are absent from the ectoderm in embryos treated with Page 20 of 51 either nickel chloride or SB203580, and identified candidates as genes down-regulated by both treatments compared to controls. Using loss of function (LOF) analysis, we tested 10 candidates and found that each is required for normal PMC positioning and skeletal patterning but not skeletogenesis, while none impacted ectodermal DV specification, as assessed by ciliary band labeling. We identified the ventrally-expressed sulfate transporter, LvSLC, as required for ventral and anterior PMC positioning and ventral/anterior skeleton formation, and show that a ventral-to-dorsal gradient of sulfated proteoglycans (sPGs) is present in control embryos and correlates spatially with SLC expression. This gradient is flattened by SLC LOF, suggesting that sPGs are an attractive cue for ventral and anterior PMCs. We also identified ventrally expressed Notch2 as required for the dorsal localization of PMCs and dorsal skeleton formation, suggesting that ventral Notch2 functions to repel PMCs dorsally. These results suggest that together, SLC and Notch2 define the initial PMC distribution. We tested this hypothesis with combined SLC and Notch2 LOF, and the results show that double morphants exhibit disrupted PMC positioning at late gastrula, which does not resolve at later timepoints, and a failure to secrete a skeleton. Double morphants possess a restricted ciliary band, indicating the ectodermal dorsal-ventral specification is intact. Together, these data identify SLC and Notch2 as specifically mediating initial dorsal-ventral PMC positioning. 34 G protein regulation of cell shape change in the early sea urchin embryo Shuster, Charles b. (New Mexico State University); Ellis, Andrea (New Mexico State University); Sepulveda, Silvia (New Mexico State University); Alvarez, Anthony (New Mexico State University); De La Rosa, Richard (New Mexico State University) The actin cytoskeleton is the primary determinant of cell shape and the driving agent of shape change in animal cells. As blastomeres of the early embryo undergo cycles of cell division, they also undergo a gradual transition from a spherical to an epithelial morphology, and we are focused on understanding the cytoskeletal dynamics underlying these processes. Using live cell probes, we now know that in addition to cortical and microvillar actin, there are cytoplasmic populations of actin in sea urchin eggs, and all of these filament populations undergo dynamic changes during the cell cycle. These actin populations are presumably regulated by Rho family GTPases, and while Rho is known to be the master regulator of contractile ring formation during cytokinesis, the roles of Rac and Cdc42 in the early embryo are less well defined. Expression of activated mutants of both Rac and Cdc42 lead to cytokinesis failure. Interestingly, activated Rac also resulted in exaggerated blastomere cohesion, suggesting that Rac activity may be involved in the spherical to epithelial shape change that occurs in later divisions. Dominant-negative mutants of Rac and Cdc42 had no effects on early cleavages, consistent with the notion that these factors are not required for cell division. However, co-expression of T17N mutants of Rac and Cdc42 led to dramatic alterations in blastomere organization, suggesting that these factors play partially redundant roles in regulating spindle orientation. Current efforts are focused on defining the molecular basis by which Rac and Cdc42 antagonize the cytokinetic apparatus, as well as explore the roles of these G proteins in the regulation of cell adhesion and cell polarity in the early embryo. 35 Three cell behaviors involved in shaping the ascidian gastrula : cell cycle duration, unequal cleavage, and oriented cell division. McDougall, Alex (Sorbonne Universities/CNRS); Dumollard, Remi (Sorbonne Universities/CNRS); Chenevert, Janet (Sorbonne Universities/CNRS); Costache, Vlad (Sorbonne Universities/CNRS); Hebras, Celine (Sorbonne Universities/CNRS) Cell cycle control mechanisms affect cell number, size and position in the ascidian embryo. Cell number is controlled by a gene regulatory network (GRN) initiated by b-catenin at two key points during embryogenesis. First, at the 16 cell stage nuclear b-catenin in endomesodermal cells causes them to Page 21 of 51 cycle faster than the ectoderm cells lacking b-catenin giving rise to the 24 cell stage. Two cell cycles later b-catenin slows down endodermal cells giving rise to a 112 cell embryo at the time of gastrulation. During this time from the 8 to the 64 cell stage two posterior vegetal cells undergo three successive rounds of unequal cleavage generating two small posterior cells at the 64 cell stage that will form the germ lineage. Here a maternal mechanism leads to the formation of a cortical structure that causes unequal cleavage by displacing the spindle from the cell center and reorienting it. In all other cells up to the gastrula stage (112 cells) the mitotic spindle is oriented parallel to the outside of the embryo. Comparing the mathematical prediction of cell division plane with actual cell division planes reveals that cells divide according to the long-axis rule (with an apical constraint), unless they display unequal cleavage. The precise topographical positioning of most blastomeres up to the gastrula stage is therefore defined by an apical long-axis rule. These three mechanisms partly explain how ascidian embryos modulate the cell cycle to generate their invariant cleavage pattern up to the gastrula stage. We have gone some way to explain the invariant cleavage pattern that was formalized by a blastomere nomenclature system developed by Conklin in 1905. 36 David Burgess 37 Functional analysis of microRNAs in development Song, Jia L. (University of Delaware); Stepicheva, Nadezda (University of Delaware); Nigam, Priya (University of Delaware) microRNAs (miRNAs) are 22-nucleotide RNAs that are expressed in both animals and plants. In animal cells, they fine tune gene expression by pairing to the 3’ untranslated region of protein coding mRNAs to repress their translation and/or induce mRNA degradation. Relatively little is known about the function of specific miRNAs in animal cells because of the difficulty in miRNA target prediction as well as the presence of miRNA families with redundant target genes/function in mammals. The majority of the miRNA gene families in Strongylocentrotus purpuratus contain a single member which makes it a tractable model to examine the function of individual miRNAs. While we have a rich knowledge on transcription factor based regulation in cell specification, virtually nothing is known about the regulatory role of miRNAs in early development. Therefore, we use the sea urchin gene regulatory network as a strong basis to fill the knowledge gap of miRNA function in developmental pathways. We integrate the regulatory roles of miRNAs into the canonical Wnt signaling pathway, which is highly conserved and essential for the specification of endoderm and mesoderm in metazoans. Using luciferase reporter constructs and site-directed mutagenesis, we found Dishevelled and β-catenin to be directly regulated by at least one shared miRNA. Blocking miRNA regulation of the β-catenin gene resulted in significant increase in β-catenin protein accumulation, increased transcript levels of Wnt responsive endodermal regulatory genes, and aberrant gut development. Results from this study elucidate the critical regulatory roles of miRNAs in early development, identify conserved miRNAs that modulate the Wnt pathway, and provide unprecedented insight into the functional role of miRNAs in the developing embryo. 38 The ectoderm-mesoderm connection and the upstream regulation of VEGF and VEGFR Ben-Tabou de-Leon, Smadar (The University of Haifa, Israel); Gildor, Tsvia (The University of Haifa, Israel) The sea urchin larval skeleton is formed by the skeletogenic mesoderm (SM) cells through epithelial to mesenchymal transition, cell migration, cell fusion and biomineralization. The communication between the ectoderm and the SM cells through VEGF signaling is necessary for the skeleton formation. According to the current model, the SM specification and the ectoderm patterning are controlled by independent regulatory mechanisms and the coordinated activation of VEGF and VEGFR are simply due Page 22 of 51 to the sequential activation of regulatory genes in each tissue. Yet, various signaling pathways and regulatory genes are active at both the ectoderm and the SM and some of them could be part of the SM-ectoderm coordination device. Here we seek to determine whether the upstream regulation of VEGF is coordinated with or independent of the upstream regulation of VEGFR. To that end, we perturb the expression of key ectodermal regulatory genes and study the effect on VEGF and VEGFR spatio-temporal expression pattern. To verify our perturbation analysis results we use VEGF recombinant GFP BAC to identify a minimal cis -regulatory region that recapitulates VEGF expression pattern. This minimal region contains binding sites of predicted ectodermal regulators. We believe that our studies will determine the mechanisms that mediate the coordinated expression of VEGF and VEGFR and illuminate the basic principles of inter-tissue communication during organ formation. 39 Late Alk4/5/7 activity is required for anterior skeletal patterning in sea urchin embryos Piacentino, Michael (Boston University); Ramachandran, Janani (Boston University); Bradham, Cynthia (Boston University) Skeletal patterning in the sea urchin embryo requires a conversation between the skeletogenic primary mesenchyme cells (PMCs) and the overlying pattern-dictating ectoderm; however, our understanding of the molecular basis for this process remains incomplete. Here, we show that TGF-ß-receptor signaling is required during gastrulation to pattern the anterior skeleton. Treatment with SB431542 (SB43), a specific inhibitor of the TGF-ß type I receptor Alk4/5/7, during gastrulation blocks anterior PMC positioning in the oral hood and the formation of the animal skeleton, but does not perturb ciliary band restriction or neural development. SB43 treatment during gastrulation does not perturb dorsal-ventral specification, but does perturb left-right axis specification, as expected. Feeding experiments show that, while Alk4/5/7 inhibition does not prevent the formation of a mouth, SB43-treated plutei display reduced feeding ability, presumably due to the loss of the anterior skeleton. Both Univin and Nodal are potential ligands for Alk4/5/7; however, Nodal is unilaterally expressed on only the right side, while Univin is bilaterally expressed in the ectoderm adjacent to the animal skeleton during the relevant time period. Our results demonstrate that Univin is necessary and sufficient for secondary skeletal patterning, consistent with the hypothesis that Univin is the relevant Alk4/5/7 ligand for anterior skeletal patterning. Taken together, our data demonstrate that Alk4/5/7 signaling during gastrulation is required to direct PMCs to the oral hood, and suggest that the relevant ligand for this signaling event is Univin. 40 Segregation of pigment and blastocoelar cells depends on the interplay between the lineage specific transcription factors ESE and GCM Molina Jiménez, Mª Dolores (Institut de Biologie Valrose (iBV), UMR7277, CNRS/UNSA); Lepage, Thierry (Institut de Biologie Valrose (iBV), UMR7277, CNRS/UNSA) The non-skeletogenic mesoderm (NSM) is initially specified by Delta/Notch signalling as a unique population of gcm-positive mesodermal precursors that will eventually differentiate into two main cell types: blastocoelar cells, which form on the ventral side and lose expression of gcm, and pigment cells, which form on the dorsal side and retain expression of gcm. This dorsal–ventral patterning depends on the antagonistic interplay between Nodal and BMP2/4 signals: Nodal signals emanating from the ventral ectoderm promote specification of blastocoelar cells and repress formation of pigment cells while dorsally, BMP2/4 counteracts the activity of Nodal, allowing pigment cell differentiation. The transcription factor Not represses gcm expression in the ventral NSM downstream of Nodal signalling, allowing expression of blastocoelar lineage specific transcription factors such as gataC, ese, and scl, and specification of blastocoelar cell fates. Here, we show that the blastocoelar cell specific transcription factor ese and the pigment cell specific transcription factor gcm appear initially coexpressed in the precursors of the immunocytes at the hatching blastula stage. Importantly, Nodal and BMP2/4 are not Page 23 of 51 required for this early ese and gcm expression but to resolve and pattern a population of cells with an initial mixed identity. By the time of delamination of the primary mesenchymal cells, segregation of ese and gcm expression and DV patterning of the NSM occurs. Ventral NSM cells retain expression of ese and lose expression of gcm, while dorsal NSM lose expression of ese and retain expression of gcm. Perturbation and overexpression experiments indicate that Ese is necessary and sufficient to promote expression of blastocoelar cells lineage specific transcription factors such as gataC, prox1 and scl on the ventral population of NSM as well as to restrict gcm expression to the dorsal population of NSM precursors. Thus, we identify Ese as an key component of the gene regulatory network responsible of blastocoelar cell specification. 41 SoxC functions in neural precursor cells in sea urchin embryo neurogenesis Wei, Zheng (NIH/NIDCR); Angerer, Lynne (NIH/NIDCR); Angerer, Robert (NIH/NIDCR) Previously we showed that Six3 was necessary for development of all nerves in the sea urchin embryo (Wei et al., 2009). The expression of Six3 begins during cleavage stages broadly throughout the embryo and then is confined to patches of contiguous cells; but it is not expressed in individual neural precursors. This suggests that Six3 is involved in establishing neuroectoderm territories but is not directly involved in initiating neural differentiation. To look for Six3-dependent genes that might execute Six3's function in neurogenesis, we carried out a microarray-based screen at the mesenchyme blastula stage when expression of proneural factor orthologs begins to appear in individual cells. Among the set of Six3-dependent transcription factors expressed in individual cells, SoxC has a unique function in neural precursor cells. By in situ hybridization, we found that SoxC is expressed in individual cells in neuroectoderm. More importantly, when SoxC was knocked down with morpholino oligos, development of all neurons was greatly reduced as is also the case in Six3 morphants. However, in a double in situ with Synaptagmin B, which is a marker for almost all neurons, no cells expressed both. This observation suggested that SoxC-expressing cells could be in an intermediate state and other factors mediate their further differenciation. We carried out a RNA-Seq screen for potential candidates. Among the affected genes, we tested Z167 and Brn1/2/4 and found that they both are partially co-expressed with SoxC and partially co-expressed with Synaptagmin B or Tph, a marker for Serotonin cells. Therefore, in sea urchin embryo, neurogenesis is a multistep process and SoxC functions in neural precursors, which is a conserved function observed in other systems. 42 Suppression of nodal expression in prospective dorsal cells of the early sea urchin embryo by the Hbox12 homeodomain regulator Cavalieri, Vincenzo (University of Palermo) Dorsal/Ventral (DV) axis formation in the sea urchin embryo depends upon the expression of nodal on the ventral side, which behaves as a DV organizing centre. However, only fuzzy clues are known as to the early symmetry-breaking steps that lead to the positioning of such an organizer. An extremely interesting candidate for this role is the hbox12 homeobox-containing gene. In Paracentrotus lividus , hbox12 expression is antecedent and complementary with respect to that of nodal , being confined in prospective dorsal cells. We show that ectopic expression of Hbox12 provokes DV abnormalities and attenuates nodal as well as nodal -dependent gene transcription. By blastomere transplantation, we also establish that DV defects arise from hbox12 misexpression in the animal hemisphere. To impair Hbox12 function we expressed ubiquitously a truncated form of the protein, encoding for the homeodomain. Such a perturbation disrupts DV axis formation by allowing ectopic expression of nodal across the embryo. Moreover, clonal loss-of-function imposed by either blastomere transplantation or gene transfer assays highlights that Hbox12 action in prospective dorsal cells is necessary for DV polarization. Remarkably, Page 24 of 51 the localized knock-down of nodal restores DV polarity of embryos lacking hbox12 function. Finally, we show that hbox12 is involved in the dorsal-specific inactivation of the p38 MAPK, which is known to be required for nodal expression. Altogether, our results indicate that Hbox12 prevents the ectopic activation of nodal transcription within the future dorsal side of the early sea urchin embryo. 43 Evolution of ectoderm-mesoderm communication during skeletal patterning in echinoid larvae Lyons, Deirdre (Duke University); Martik, Megan (Duke University); Kimura, Julian (Duke University); McClay, David (Duke University) The echinoid larval endoskeleton is a classic model for studies of cell differentiation and morphogenesis. It is also a model for the evolution of embryonic patterning, since specific parts of the skeleton can vary between species. Two of the most variable elements are the recurrent and posterior connecting rods. The developmental basis of this variation is not well understood. By comparing the sea urchin Lytechinus variegatus , which forms a small recurrent rod and no posterior connecting rod, with the sand dollar Mellita quinquiesperforata , which forms both, we found that the difference in their skeletal patterning is already obvious in the arrangement of primary mesenchyme cells (PMCs) at gastrula stages. Mellita embryos possess an antero-dorsal chain of PMCs between the tips of the longitudinal strands, which emanate from the ventro-lateral clusters. These PMCs contribute to the recurrent rod and the anastomosed posterior connecting rod. No antero-dorsal chain exists in Lytechinus , but at prism stages it is from an analogous location that the recurrent rods form from a branching event at the tip of each longitudinal strand. Because the position of the PMCs is dictated by ectodermal patterning in sea urchins, we investigated the role of ectodermal patterning in Mellita PMC arrangement. We found that Nodal signaling controls the position of the antero-dorsal chain in the oral/aboral axis. Ectodermal signals that control the spatiotemporal pattern of underlying PMCs have diverged between these two echinoids. These data provide the framework for studies in both species that address the details of these two patterning systems at the molecular level. 44 Experimental approach to divergence in test organization between euechinoid and cidaroid sea urchins Gao, Feng (California Institute of Technology); Erkenbrack, Eric (California Institute of Technology); Petsios, Elizabeth (University of Southern California); Thompson, Jeffrey (University of Southern California); Bottjer, David (University of Southern California); Davidson, Eric (California Institute of Technology) The two extant crown groups of echinoids are the subclasses Cidaroidea and Euechinoidea, which diverged around P/T boundary and differs strikingly in several aspects of skeletal morphogenesis. Such divergence thus represents a compelling opportunity for understanding the evolutionary mechanisms of specific body plan innovations in echinoderms. Our initial focus is on the difference of their test pattering, a major phylogenetic character as revealing from their development and fossil records. Embryos from Strongylocentrotus purpuratus (Euechinoid) and Eucidaris tribuloides (Cidaroid), were cultured in the lab to the stage six weeks after metamorphosis, and samples were collected at different time points. The whole animal and extracted skeletal elements were scanned under SEM and micro-CT to know how their tests are built differently in these two species by addition of successive rows of body plates at the start of adult skeletogenesis. The difference on their test patterning was first visible from one week after metamorphosis with compound plates in Sp vs simple plates in Et from the ambulacral region while no discernible difference from the interambulacral region. WMISH was done on genes from the gene regulatory network underlying the development of the skeletogenic lineage of Echinoderm to find what accounts for the clade–specific differences in the spatial deployment of this GRN on the ambulacral region in Sp and Et. Gene regulatory analysis and synthetic experimental evolution will be Page 25 of 51 done in the future, and our final goal is to understand the mechanistic origin of the Euechonoid compound platting at GRN level. 45 Evolution of pancreatic cell types: insights from the sea urchin Strongylocentrotus purpuratus Perillo, Margherita (Stazione Zoologica Anton Dohrn di Napoli, Naples, Italy); Arnone, Maria Ina (Stazione Zoologica Anton Dohrn di Napoli, Naples, Italy) The proper pancreas of vertebrates is a gland composed essentially of digestive enzymes-producing acinar cells and hormone-producing endocrine cells. Although the pancreatic hormone insulin is well characterized in vertebrates, little information is reported about insulin-like peptides (ILPs) in non-chordate deuterostomes. In order to fill this critical gap in pancreas evolution, our aim is to characterize the homologues of pancreatic cell types and ILPs in the embryos and larvae of Strongylocentrotus purpuratus, so far one of the best non-chordate deuterostome for evolutionary comparison studies. To this end, the spatial expression of ILPs found in the sea urchin genome (two paralogues named SpILP1 and SpILP2) and their predicted receptor (SpInsr) has been extensively studied. Remarkably, SpILP1 has been found to be localized in a group of cells of the larval gut in a feeding-dependent fashion. Moreover, to characterize homologues of pancreatic cell types, we characterized the spatial expression of the orthologues of pancreatic transcriptional factors (SpNgn, SpNeuroD, SpIsl, SpHnf1, SpPtf1a, SpMist1) and exocrine pancreas terminal differentiation genes (SpCpa2L, SpPnlp, SpAmy3), which expression increases after feeding. Notably, perturbation analysis experiments also demonstrated that in the sea urchin the link between SpHnf1, SpPtf1a and the pancreatic digestive enzymes is conserved, thus allowing us identifying an acinar-like cell type in the upper stomach of the sea urchin larva. In addition, the already identified SpLox&SpBrn1/2/4+ cells have been further characterized and we found that it is a neurosecretory cell type that produces a novel neuropeptide. Furthermore, comparing the above outcome together with available data in other animal models, we propose a model of pancreatic cell types evolution across metazoan. Concluding, comparison of the ILPs characterized in the sea urchin with proteins from other bilaterians were helpful to carry out a phylogenetic study aimed to explain how molecules of the insulin family evolved. 46 Regulatory gene use within the gastrulating sea urchin Valencia, Jon E. (Caltech) Our goal is to produce a regulatory gene expression dataset that will aid in the systems-level understanding of cell-type specification and organ formation in the post-gastrula embryo. A complete understanding of the regulatory genes that comprise the regulatory states formed during development is the first step in building GRNs. Previous regulatory gene surveys analyzed temporal and spatial expression up to the onset early gastrulation. We present a complete spatio-temporal characterization (from 36 to 72 hours post fertilization) of gene expression patterns for all known and predicted transcription factors genes in the sea urchin genome. The genome contains 368 transcription factors genes (non zinc fingers and identified zinc fingers) and only 284 of those are significantly expressed during the hours of gastrulation as determined to be relevant by RNA-seq data. We have analyzed gene expression patterns by whole-mount in-situ hybridization for 80% of the regulatory genes expressed at four time-points during and post gastrulation and have annotated these patterns into the six major domains of the developing sea urchin. Interestingly, our analysis of this dataset has revealed that the non-skeletal mesoderm, endoderm, oral ectoderm and apical ectoderm each expresses around 50% of the regulatory genes considered, whereas skeletal mesoderm and aboral ectoderm each expresses less than 20%. These results not only show an enormous regulatory complexity but also that most regulatory genes are expressed in multiple domains. Additionally, our results offer many insights into the global use of regulatory genes expressed during sea urchin development. Page 26 of 51 47 Delta-Notch signaling and HesC mediate the spatial confinement of the skeletogenic-specific regulatory gene alx1 to micromere-descendants in Eucidaris tribuloides Erkenbrack, Eric (Caltech); Davidson, Eric (Caltech) The early restriction of skeletogenic regulatory genes—namely alx1 , delta , ets1 , tbrain —to micromere-descendants via the Pmar1-HesC double-negative gate is a widespread piece of GRN circuitry in euechinoids, having been described in at least three euechinoid Orders. Evidence for this specific circuit is absent outside of euechinoids, raising the evolutionary question of how and when it was installed. We undertook GRN analysis of skeletogenic regulatory genes in a cidaroid sea urchin, Eucidaris tribuloides ( Et ), as it is a representative of a distantly-related clade from euechinoids (~275 mya) and manifests several significant developmental differences with euechinoids. Repeated BLAST searches of genomic and transcriptomic data sets of Et yielded no significant hits for pmar1 , supporting the supposition that pmar1 is a euechinoid novelty. In Et , delta is first zygotically expressed in micromere-descendants at early blastula stage, rather than during cleavage as in euechinoids. HesC is maternal and is zygotically transcribed shortly after delta transcription begins. However, rather than being expressed everywhere but the skeletogenic micromeres as in euechinoids, this repressor is expressed exclusively in mesodermal cells immediately surrounding micromere-descendants. Alx1 is zygotically expressed exclusively in micromere-descendants; while ets1 and tbrain are general mesodermal drivers--the latter being maternal. We found that the zygotic expression of hesc is Delta-dependent. The absence of HesC (and also of Delta presentation) causes the alx1 domain to expand to surrounding mesoderm cells, but not to the whole embryo as in euechinoids. Thus the spatial confinement of alx1 to micromere-descendants in Et is Delta-Notch dependent. Evolutionarily, HesC-mediated repression of alx1 is likely an ancestral linkage in the echinoid skeletogenic GRN. The global function of preventing skeletogenesis by HesC in euechinoids is not required in Et ; rather its only skeletogenic role is to separate skeletogenic from non-skeletogenic mesodermal fates during embryogenesis. 48 Modularity in DNA Binding Preference of a Tbrain Transcription Factor May Allow for More Versatile Transcriptional Responses and Increased Evolvability Cheatle Jarvela, Alys (Carnegie Mellon University) In the sea star (Pm), Tbrain (Tbr), a t-box transcription factor, carries out a variety of ancestral roles in the endomesoderm and ectoderm. However, in the sea urchin (Sp), Tbr has a singular and well-defined role in the skeletogentic gene regulatory network. The DNA binding regions of these proteins contain differences in critical DNA-contacting amino acids, suggesting a protein level change could be responsible, but it is thought that this type of change must be incredibly rare owing to the potentially lethal pleiotropic effects of altering a multifunctional protein. However, novel techniques have allowed for far more sensitive assays of transcription factor function. One such technique, Protein-Binding Microarrays, has indicated that DNA binding is more complex than originally indicated, in that the same transcription factor can recognize multiple binding sites, and only a subset of these might be conserved among closely related paralogs (Badis and Bulyk 2009). Here, we first demonstrate that this type of complexity also applies to orthologous transcription factors. While both Tbr orthologs recognize the same primary motif, only PmTbr also has a secondary binding motif. While affinity for the primary binding site is conserved, affinity for the secondary binding motif is more evolutionarily labile. We have verified the effects of binding specificity and affinity changes in vivo using a dual Otxb1/2 CRM reporter system. Our in vivo assays demonstrate that differences in transcriptional responses governed by Page 27 of 51 primary vs. secondary sites may allow for greater evolution in timing of regulatory control. This uncovers a layer of transcription factor binding divergence that could exist for many pairs of orthologs. We hypothesize that division of transcriptional functions between multiple binding sites may allow orthologs to evolve new secondary binding preferences rapidly, as the conserved primary site reduces pleiotropic effects. 49 Germ cells Rock! Wessel, Gary M. (Brown University) They really do.... 50 Michael Whitaker 51 Translatome analysis following fertilization Morales, Julia (equipe TCCD, UMR8227 CNRS-UPMC, Station Biologique de Roscoff); Chassé, Héloïse (equipe TCCD, UMR8227 CNRS-UPMC, Station Biologique de Roscoff); Boulben, Sandrine (equipe TCCD, UMR8227 CNRS-UPMC, Station Biologique de Roscoff); Corre, Erwann (ABiMS, FR2424 CNRS-UPMC, Station Biologique de Roscoff); Le Corguillé, Gildas (ABiMS, FR2424 CNRS-UPMC, Station Biologique de Roscoff); Glippa, Virginie (equipe TCCD, UMR8227 CNRS-UPMC, Station Biologique de Roscoff); Coson, Bertrand (equipe TCCD, UMR8227 CNRS-UPMC, Station Biologique de Roscoff); Belle, Robert (equipe TCCD, UMR8227 CNRS-UPMC, Station Biologique de Roscoff); Mulner-Lorillon, Odile (equipe TCCD, UMR8227 CNRS-UPMC, Station Biologique de Roscoff); Cormier, Patrick (equipe TCCD, UMR8227 CNRS-UPMC, Station Biologique de Roscoff) Half of the variation in proteins concentration in a cell can be attributed to differences in translation rates as determined by global quantification of gene expression. Therefore, protein synthesis represents an import step in gene expression regulation. Early embryogenesis is highly dependent on translational regulatory cascades, and relies on the use of maternally stored mRNA in the cytoplasm. Sea urchin embryos offer an elegant model to elucidate the network of actors involved in translational control during early development. Protein synthesis, low in unfertilized eggs and stimulated rapidly following fertilization, is necessary for the onset of the first cell divisions. We have shown previously that translational machinery is highly regulated and activated at fertilization, acting upon translation initiation and elongation steps. Translatome, a subset of the transcriptome comprising polysomes-associated mRNAs, corresponds to the totality of mRNAs translated into proteins thus reflecting the functional readout of the genome. A complete examination of translatome is made possible by carrying out polysome profiling coupled with high-throughput sequencing technologies. We have undertaken the analysis of the translatome following fertilization and showed selective recruitment of mRNAs encoding translation factors and RNA binding proteins. From these data on translatome analysis and our previous results on translation machinery activation at fertilization, we suggest that a translational regulatory cascade is important for the first few hours of development of the embryo. 52 Gene regulatory networks in the real world: stress, robustness, and evolution Wray, Gregory (Duke University) Development produces a reproductively competent adult despite a wide range of genetic and environmental perturbations. At the same time, development retains the flexibility to to evolve in response to environmental change. We are actively investigating how sea urchins balance these conflicting demands on development gene regulatory networks. We compared the effects of three kinds of real world perturbations: natural genetic variation and two environmental stressors linked to global warming, pH and temperature. Both projected environmental stressors have measurable impacts on Page 28 of 51 developmental phenotypes and on proxies for fitness. Nonetheless, development is able to buffer both environmental stressors at least as well as they are able to buffer the consequences of natural genetic variation. Overall, our results indicate that early development in sea urchins is highly robust to genetic and environmental perturbation, but that specific gene interactions are more sensitive and may allow adaptive changes in organismal traits. 53 A prelude to the present: Paleontological perspectives on 450 million years of echinoid evolution Thompson, Jeffrey R. (University of Southern California); Petsios, Elizabeth (University of Southern California); Bottjer, David (University of Southern California) Echinoids are a morphologically diverse group of echinoderms with a robust fossil record dating back to the Upper Ordovician period, about 450 million years ago. The fossil record of echinoids provides valuable data to both paleontologists and neontologists interested in evolutionary processes. But what data are available to neontologists through study of the fossil record? The fossil record of the Echinoidea allows for the testing of evolutionary hypotheses and provides insight into how large-scale events such as global climate and sea level change govern evolution and diversification processes. Following the Permian-Triassic mass extinction, 252 million years ago, echinoid diversity underwent a bottleneck and concomitant reduction in morphological disparity. The surviving bauplans, with only two columns of plates in each interambulacral and ambulacral column became the template for all post-Paleozoic, and thus recent, echinoids. Using a paleogenomic approach, it is possible to place changes in the GRN and genome within this context of evolutionary time. The fossil record provides the only evidence available to directly observe trait reversals, especially in relation to extinct stem-groups and also provides observable evidence for the morphologic manifestation of changes in the skeletogenic gene regulatory network circuitry. Fossil and molecular developmental approaches are now being used to understand the divergence between cidaroids and the euechinoids, the two clades that comprise all post-Paleozoic echinoids. Thus far, fossil data has elucidated better estimates for the timing of this divergence and for the timing of the first appearance of ordinal and family level traits, and the associated regulatory circuitry. 54 Pattern and process during gut morphogenesis: an evolutionary perspective Arnone, M. Ina (Stazione Zoologica Anton Dohrn) It can be argued that one of the first developmental patterning systems to evolve was the molecular network that orchestrates the formation of the digestive system thus releasing multi-cellular organisms from body size constraints and allowing the further evolution of highly specialized internal structures. The antero-posterior patterning of the embryonic gut represents one of the most intriguing biological processes in development. A dynamic control of gene transcription regulation and cell movement is perfectly orchestrated shaping a functional gut in distinct specialized parts. Two ParaHox genes, Xlox and Cdx, play key roles in vertebrate and sea urchin gut patterning through molecular mechanisms that are still mostly unclear. We combined functional analysis methodologies with high resolution imaging and RNA-seq to investigate Xlox and Cdx regulation and function in the sea urchin embryo. We revealed part of the regulatory machinery responsible for the onset of Cdx transcription, uncovered a Wnt10 signal mediating Xlox repression in the intestinal cells and provided evidence of Xlox control of the stomach differentiation. A role for the retinoic acid mediated signalling to initiate this process was also uncovered. Our findings offer a novel mechanistic explanation of how the control of transcription is linked to cell differentiation and morphogenesis for the development of the sea urchin larval gut. A comparison with vertebrates showed a striking conservation of topology of gene expression and Page 29 of 51 signalling events between sea urchin and mouse, thus revealing some key aspects of deep homology that are most probably shared by all bilaterian guts. Preliminary analyses, comparing sea urchin and sea star gut patterning, suggest that this conservation does not extended to the level of gene interactions. 55 Differentiation of immunocytes and the emergence of the larval immune system Rast, Jonathan (University of Toronto); Buckley, Katherine (University of Toronto); Ho, Eric (University of Toronto); Schrankel, Catherine (University of Toronto); Wang, Guizhi (University of Toronto) Animal immunity is an organism-wide phenomenon involving specialized immune cells, barrier epithelia and other tissues. In the gut associated immune system of Bilateria, interactions among the lumenal microbiota, the gut epithelium, dedicated immunocytes and peripheral tissues have broad effects on throughout the host. The feeding larva of the purple sea urchin provides a deuterostome model that is well-suited to investigate these processes. Larvae are morphologically simple and transparent, which allows organism-wide imaging at single-cell resolution. Upon exposure to Vibrio diazotrophicus, several classes of specialized immune cells exhibit stereotypic changes in morphology, undergo migration to the gut epithelium and specific interactions among themselves. These behavioral changes coincide with changes in gene expression in the gut. Aspects of this response are synchronous among individuals and reproducible. Transcriptome data from larvae exposed to bacteria at several time points and treatments provide a system-wide measure of transcriptional changes. Notably, expression of several IL-17 homologs is quickly induced in the gut epithelium. These signals are tightly regulated, expressed exclusively in presence of bacteria and are the most strongly upregulated in the genome. Deletion analysis of IL-17 transgenes localizes response and attenuation elements. Antisense perturbation of IL-17 receptors identifies candidate downstream genes. Later in the response, transcription levels change throughout the organism for genes encoding transcription factors (e.g., NfkB, PU.1, and Irf), signaling molecules (e.g., TNF and Mif), pattern recognition receptors (e.g., PGRPs) and immune effectors (e.g., 185/333 genes). The molecular complexity of this inflammatory response within the context of the simple morphology of the sea urchin larva provides an ideal system in which to characterize the fundamental programs of immune response mediated by the gut epithelium. 56 A GRN for neurogenesis in the sea star P. miniata Hinman, Veronica (Carnegie Mellon U) Nervous systems are centralized in various taxa, distributed throughout the animal tree of life. It has been of great interest to understand how such complex structures can evolve from diffuse neural nets and whether the transition had a single origin. The evolution of the extraordinary complex vertebrate nervous system has been especially puzzling to understand, in part because their sister taxa, the hemichordates and echinoderms have very simple nervous systems. Although sea stars had featured prominently in early hypotheses of vertebrate neural origins, an increasing understanding of molecular mechanism of development have suggested alternative hypotheses that imply that the sea star, and echinoderms generally have derived nervous systems. We have developed extensive gene regulatory networks that explain how the sea star Patiria miniata larval nervous system is specified and patterned. This work reveals an extraordinary similarity in regulatory mechanism of development between these echinoderms and vertebrates. More significantly, these analyses demonstrate how the modular nature of these regulatory networks, and in particular the independence between patterning and specification, can explain how nervous systems patterning can readily evolve. 57 Spiralian embryogenesis at your fingertips: the quest for an early annelid GRN Schneider, Stephan (Iowa State University); Pruitt, Margaret (Iowa State University); Bastin, Benjamin (Iowa State University); Chou, Hsien-chao (Iowa State University) Page 30 of 51 Although conserved features among developmental gene regulatory networks (GRNs) between various deuterostome embryos are emerging, a comparison to protostomes remains difficult due to the highly derived nature of early embryogenesis and the genomes in the protostome models C. elegans and Drosophila (Ecdysozoa). To fill this gap we focus on early developmental mechanisms in the protostome Platynereis dumerilii (Annelida, Lophotrochozoa). Platynereis has retained a more ancestral gene set without the extensive gene loss and gain observed in the other protostome models. In addition, early development of Platynereis is attractive as it exhibits a series of invariant, stereotypic asymmetric cell divisions, allowing individual embryonic cells to be recognized by size and position. Furthermore, Platynereis exhibits a global reiterative beta-catenin mediated cell fate specification mechanism in early development that appears to convey lineage-specific binary cell fate decisions. To gain insights into the early embryonic GRNs, and the contribution of reiterative beta-catenin asymmetries to specify cell fates, we deployed a variety of RNA-seq based approaches. Comparing normal and manipulated embryos we have identified embryonic transcripts from the zygote through the mid gastrula (1 cell stage through ~330 cell stage) that include developmental regulators of the beta-catenin pathway. We have subsequently mapped the expression of developmental regulators including beta-catenin pathway components (ligands, receptors, intracellular components) into distinct cell lineages. Our approaches capture stage-specific transcriptional snapshots of Platynereis embryos, and thereby provide the first comprehensive view of temporal inputs and outputs into annelid GRNs that utilize a spiral-mode of cell divisions to segregate cell fates. 58 Developmental plasticity: A broad utilization of germ line molecules in multipotent cells of the sea urchin Yajima, Mamiko (Brown University); Wessel, Gary (Brown University) Echinoderms are classically known for having remarkable regenerative capabilities. Adults can regenerate entire segments such as arms, internal organs and even gonads, and embryos maintain multipotency until late in their development: Cells from other lineages can transfate and compensate for a missing part of the embryo. The mechanisms that regulate this amazing plasticity of echinoderm cells are not yet clear, but we recently found that a transient use of germ line molecule may be taking a critical role in facilitating cellular multi-potentiality in the multipotent cells of the sea urchin embryo. In this talk, we focus on a conserved germ line molecule, Vasa, and demonstrate how it is transiently expressed, control cell cycle progression, and further regulate general protein synthesis necessary for the embryogenesis, developmental reprogramming and/or regeneration. From the results showed in this talk, we propose Vasa functions broadly in mRNA regulation to enable proper distribution and efficient translation of mRNAs that contribute to developmental plasticity of the sea urchin embryo. 59 Maintenance of the anterior neurogenic ectoderm in the sea urchin embryo Yaguchi, Junko (University of Tsukuba); Yaguchi, Shunsuke (University of Tsukuba); Yamazaki, Atsuko (University of Tsukuba); Yamamoto, Akane (University of Tsukuba); Inaba, Kazuo (University of Tsukuba) In normal sea urchin embryos, the anterior neurogenic ectoderm is restricted to the anterior end although almost all of cell fate will be specified as this region in pre-signaling condition. Previous works suggested that a sequence of canonical and non-canonical Wnt signal-dependent mechanisms regulate the restriction of the neurogenic ectoderm, but it is still unclear how the restricted neurogenic ectoderm is stably maintained during the early embryogenesis. Here we show that a maternally expressed transcription factor, Meis, is required for the maintenance of the anterior neurogenic ectoderm. Cis-regulatory analysis of FoxQ2, a transcription factor essential for the formation of anterior neurogenic Page 31 of 51 ectoderm, revealed an important module for the maintenance of its expression. Among transcription factors that might bind to the region, we focused on Meis homeodomain protein in this study. When we block the translation of Meis by morpholino anti-sense oligonucleotides, the expression of foxQ2 was not maintained after hatching nevertheless the initial expression of foxQ2 was not affected. As a consequence, the morphant lost the anterior neurogenic ectoderm and serotonergic neurons. These results indicated that Meis is required for the maintenance of anterior neurogenic ectoderm. In addition, the expression pattern of univin, a lateral ectoderm marker, and PMC pattern shift towards more anterior side in Meis morphants than those in control embryos, indicating that Meis is also required for the precise anteroposterior patterning of the sea urchin embryo. 60 Coelomocytes and the Sea Urchin Immune System Smith, L Courtney (George Washington University) Annotation of the sea urchin genome identified and highlighted the complexity of the innate immune system in this invertebrate, which includes several large families of immune response genes. The coelomocytes are the primary mediators of this system in the adult sea urchin and are characterized as phagocytes, spherule cells, and vibratile cells. There are four morphotypes of phagocytes including polygonal, discoidal and small phagocytes, plus a newly identified type. Only the phagocytes express immune genes such as SpC3, SpBf, SpNFkB, SpTLR, and Sp185/333. The Sp185/333 gene family encodes a wide array of highly diverse but structurally similar Sp185/333 proteins that have core functions in the immune system. One recombinant (rSp0032) shows specific binding to Vibrio and yeast, but not to Bacillus, whereas separated sub-regions of rSp0032 show deregulated binding. rSp0032 binding to bacteria and yeast is mediated by LPS, flagellin and glucan, but not by peptidoglycan. Binding to LPS is specific with high affinity. rSp0032 reduces the growth rate of E. coli but not Vibrio, and does not induce phagocytosis. On the other hand, native Sp185/333 proteins are secreted from phagocytes, bind to and opsonize bacteria, retard the growth of most bacteria, and induce phagocytosis. These results suggest that the various Sp185/333 isoforms have a range of functions and that they act synergistically in the coelomic fluid. We analyzed the Sp185/333 gene expression in single phagocytes using single cell RT-PCR and amplicon sequencing. Surprisingly, individual phagocytes express a single gene of the ~50 member gene family. This intriguing result infers a much more complex and sophisticated regulatory system for controlling Sp185/333 gene expression than has been imagined previously. 61 Shifts in the Expression of Developmental Regulatory Genes Involved in the Evolution of a Novel Life History Difference Wygoda, Jennifer (Duke University); Byrne, Maria (University of Sydney); McClay, David (Duke University); Wray, Gregory (Duke University) Developmental mode can influence dispersal, gene flow, speciation and extinction rates in marine taxa and thus can have important consequences for micro- and macroevolutionary processes. While the ancestral developmental mode of sea urchins is indirect through a feeding larval stage (planktotrophic), non-feeding development (lecithotrophic) has evolved independently multiple times. In order to identify evolutionary changes in gene expression underlying this ecologically significant shift in life history, we used Illumina RNA-seq to measure expression dynamics across development in three sea urchin species: the lecithotroph Heliocidaris erythrogramma, the closely related planktotroph Heliocidaris tuberculata (3 myr), and an out-group planktotroph Lytechinus variegatus (50 myr). Our analyses draw on a well-characterized developmental gene regulatory network (GRN) in sea urchins to understand how the ancestral developmental program was altered during the evolution of lecithotrophic development. Page 32 of 51 Our results suggest that changes in gene expression profiles were more numerous during the evolution of lecithotrophy than during the persistence of planktotrophy, and this contrast is even stronger when only GRN genes are considered. We found evidence for both conservation and divergence of GRN linkages in H. erythrogramma, as well as significant changes in the expression of genes with known roles in patterning the larval skeleton and gut, which are greatly modified in lecithotrophs. Collectively, these results indicate that the transition from planktotrophic to lecithotrophic development involved a surprising number of changes to key developmental processes over a short evolutionary timescale. 62 The Developmental Transcriptome of the Ophiuroid Amphiura filiformis Dylus, David V. (University College London); Oliveri, Paola (Unviersity College London) Ophiuroids form a larval skeleton similar to echinoids and represent an evolutionarily interesting class. Unfortunately, so far sequence resources were limited for this group of animals. With recent advances in high throughput sequencing it is now possible to create cheaper and faster sequence resources for non-model organisms. Moreover, the development of new graph-theoretical software approaches enables to easily assemble sequences without prior genome availability. These facts allowed us to perform mRNA sequencing on key developmental stages (Cleavage, Blastula, Mesenchyme Blastula and Gastrula) for the brittle star Amphiura filiformis without the need of a reference genome. The sequencing was carried out on the Illumina HiSEQ 2500 platform using 2 lanes with 100bp paired end reads resulting in ~700 million reads. In order to perform a de novo transcriptome assembly, we used digital normalization followed by assembly with the trinity software package. Using all available reads our reference transcriptome resulted in ~600.000 sequences (N50: 1094). In these sequences, we found ~15.000 transcripts with high similarity to other echinoderm genes. These include 80% of transcription factors present in S. purpuratus. When re-aligning the reads for time-course expression levels we found a high correlation between our QPCR data and the transcriptome. Focusing on 231 larval skeletogenic genes identified in sea urchin, only 171 could be recognized in A. filiformis, including a cohort of genes that could not be found when conducting a similar analysis with Patiria miniata genomic sequences. Since asteroids do not form larval skeleton, this suggests the existence of a unique set of genes utilized during larval skeletogenesis in echinoids and ophiuroids. 63 Sequencing Echinoderm Genomes Cameron, R. Andrew (California Institute of Technology) The sequencing enterprise for echinoderm genomes is now changing directions as the efforts of large sequencing centers wanes while that of individual laboratories increases. In light of these changes, I will present a synopsis of the present work to decode echinoderm genomes. I will describe the breadth and nature of the various genome and transcriptome projects available in public sites and some in preparation. I will compare the quality of the various datasets and how they can be used. And lastly, I will discuss future directions for our community efforts in genome and transcriptome sequencing and annotation. 64 GRIP-Seq: Genome-wide Regulatory element Immunoprecipitation coupled with Next Generation Sequencing Tulin, Sarah L. (MBL); Bocconcelli, Carlo (Falmouth Academy); Smith, Joel (MBL) We describe a new method for identifying cis -regulatory elements. Our protocol combines elements of chromatin conformation capture (e.g., 3C, 4C), chromatin immunoprecipitation, and paired-end high-throughput sequencing. By modifying these existing methods, we enrich for active cis -regulatory elements across the genome. Our pioneer dataset, available to the community, derives from S. Page 33 of 51 purpuatus embryos collected at 24 hours post fertilization. We benchmark our results against independent findings, i.e., rigorously tested cis- elements previously identified. We find good congruence, and in particular we find a strong signal-to-noise ratio for known distal elements, marking an improvement over existing methods. A second goal of this new method is to identify which transcription start site corresponds with which element: on this score, our findings show only a modest signal, though it is too early to say how our protocol might be adjusted to yield more reliable results. In conclusion, we report a promising protocol for the genome-wide identification of active regulatory elements, a rate-limiting step in determining comprehensive, predictive network models. 65 Experimental measurement of embryonic regulatory states Nam, Jongmin (Rutgers University-Camden); Erkenbrack, Eric M. (Caltech) Regulatory state is defined as the total set of active transcription factors presented in a given cell. As an expansion of the original concept, embryonic regulatory state is defined as the sum of regulatory activities in an entire set of cells in a given embryo. Because regulatory activities of a given set of transcription factors is determined by the information present in cis-regulatory modules (CRMs), one can use a large number of CRM::reporter constructs as a reading device for embryonic regulatory states. In this study, we used a set of 120 CRMs isolated from 42 sea urchin regulatory genes to quantitatively measure regulatory states in developing embryos of the purple sea urchin, the pencil sea urchin, and the zebrafish. We will present our findings on the developmental and evolutionary relationships of the regulatory states measured in the three species. 66 PARtisan Development Moorhouse, Kathleen (Boston College); Gudejko, Heather (Boston College); Burgess, David (Boston College) Establishment and maintenance of cell polarity has become an increasingly interesting biological question in a diversity of cell types and has been found to play a role in variety of biological functions. Previously, it was thought that the sea urchin embryo remained relatively unpolarized until the first asymmetric division at the 16cell stage of development. However, there is mounting evidence to suggest that polarity is established much earlier. We analyzed roles of the cell polarity regulators, the PAR complex proteins, and how their disruption in early development affects later developmental milestones such as blastula and gastrula formation. We found that PAR6 along with aPKC and CDC42 localize to the apical cortex (free surface) as early as the 2-cell stage of development and this localization is retained through the gastrula stage. Uniquely, PAR1 also colocalizes with these apical markers through the gastrula stage, despite the formation of a polarized epithelium. Additionally, PAR1 was found to be in complex with aPKC, but not PAR6, during these developmental stages. PAR6, aPKC, and CDC42 are anchored in the apical cortex by myosin assembly. Myosin assembly was also found to be necessary to maintain proper PAR6 localization through subsequent cleavage divisions. Interference with myosin assembly prevented the embryos from reaching the blastula stage, while transient disruptions of either actin or microtubules did not have this effect. Additionally, inhibition of myosin assembly effected actin, but not myosin, localization. These observations suggest that disruptions of the polarity complex in the early embryo can have a significant impact on the ability of the embryo to reach later critical stages in development. 67 Germ Cell Associated Gene Expression is Regulated by Nodal Signaling in the Sea Star Patiria Miniata Fresques, Tara (Brown University, Wessel Lab); Zazueta, Vanesa (Brown University, Wessel Lab); Reich, Adrian (Brown University, Wessel Lab); Wessel, Gary (Brown University, Wessel Lab) Page 34 of 51 In many animals, germ cells are specified by cell-to-cell signaling events. In some of these animals, conserved germ cell markers accumulate in broad embryonic domains in early development. Successive rounds of restriction of the germ cell markers to smaller and smaller embryonic domains result through development. In these animals it is unknown which signaling pathways are involved in restricting germ cell markers to increasingly smaller embryonic domains. We noticed that in the development of the sea star P. miniata , the germ cell factor Vasa similarly becomes sequentially restricted to smaller domains, and becomes asymmetrically restricted in regards to the Dorsal/Ventral and Left/Right axis. We hypothesize that the conserved Dorsal/Ventral and Left/Right signaling pathway ligand, Nodal, is involved in restricting germ cell factors in the sea star, and thereby instructs the other cells to become the germ line. We performed RNA in situ localizations with the known germ cell marker Vasa and known Nodal signaling pathways members to test if they become asymmetrically expressed in opposite embryonic domains at the same time. We also cultured embryos in a pharmacological inhibitor designed to inhibit Nodal signaling to test if disrupting Nodal signaling causes aberrant accumulation of the germ cell marker Vasa. We found that Vasa and Nodal become asymmetry expressed at the same time in development and that disruption of the Nodal signaling pathway causes ectopic accumulation of Vasa. These preliminary data suggest Nodal is required for repression of the germ cell fate, and the germ cell mRNAs, in the ventral and right side of the embryo and identifies the first inhibitory signaling ligand that may be involved in the inductive specification of germ cells via a sequential restriction mechanism. 68 Hedgehog Signaling Requires Motile Cilia in the Sea Urchin Warner, Jacob (Duke University); Morris, Robert L. (Wheaton College); McCarthy, Ali (Wheaton College); McClay, David (Duke University) A relatively small number of signaling pathways govern the early patterning processes of metazoan development. The architectural changes over time to these signaling pathways offer unique insights into their evolution. In the case of Hedgehog (Hh) signaling, two very divergent mechanisms of pathway transduction have evolved. In vertebrates, signaling relies on trafficking of Hh pathway components to nonmotile specialized primary cilia. In contrast, protostomes do not use cilia of any kind for Hh signal transduction. How these divergent lineages adapted such dramatically different ways of activating the signaling pathway is an unanswered question. Here, we present evidence that in the sea urchin, a basal deuterostome, motile cilia are required for embryonic Hh signal transduction, and the Hh receptor Smoothened (Smo) localizes to cilia during active Hh signaling. This is the first evidence that Hh signaling requires motile cilia and the first case of an organism requiring cilia outside of the vertebrate lineage. 69 Tissue homeostasis, regeneration and negligible senescence: insight from the sea urchin Bodnar, Andrea (Bermuda Institute of Ocean Sciences); Lortie, Mae (Bermuda Institute of Ocean Sciences); Du, Colin (Bermuda Institute of Ocean Sciences); Parsons, Rachel (Bermuda Institute of Ocean Sciences); Coffman, James (Mount Desert Island Biological Laboratory) Sea urchins exhibit a very different life history from humans and short-lived model animals and therefore provide the opportunity to gain new insight into the complex process of aging. Sea urchins grow indeterminately, regenerate damaged appendages, reproduce throughout their lifespan and some species show no increase in mortality rate at advanced ages. Further, different species of sea urchins have very different reported lifespans, thus providing a unique model to investigate mechanisms underlying both lifespan determination and negligible senescence. Studies to date have demonstrated maintenance of telomeres, maintenance of antioxidant and proteasome enzyme activities and little accumulation of oxidative cellular damage with age in tissues of sea urchin species with different Page 35 of 51 lifespans. Gene expression studies using Strongylocentrotus purpuratus indicate that key cellular pathways involved in energy metabolism, protein homeostasis and tissue regeneration are maintained with age. Quantitative analyses of cell proliferation (BrdU incorporation) and apoptosis (assessed by TUNEL and the Apo ssDNA™ assays) in sea urchin tissues (muscle, nerve, esophagus and coelomocytes) indicates a low level of tissue renewal that is maintained with age in species with different lifespans ( S. franciscanus , S. purpuratus and Lytechinus variegatus ). Expression of genes involved in cell proliferation and/or pluripotency (i.e. pcna , tert , soxB1 , oct1/2 , myc , vasa , piwil2 ) is maintained with age in these tissues. Regenerative capacity, assessed by measuring the regrowth of amputated tube feet and spines in L. variegatus , is maintained with age. We postulate that long-term maintenance of mechanisms that sustain tissue homeostasis and regenerative capacity are essential for indeterminate growth and negligible senescence and an understanding of these mechanisms may reveal effective strategies to prevent the degenerative decline with age. 70 Deuterostome origins of gastric pH regulation Stumpp, Meike (Academia Sinica (ICOB)); Hu, Marian Y. (Academia Sinica (ICOB)); Tseng, Yung-Che (National Taiwan Normal University); Su, Yi-Hsien (Academia Sinica (ICOB)); Yu, Jr-Kai (Academia Sinica (ICOB)); Hwang, Pung-Pung (Academia Sinica (ICOB)) The highly acidic gastric environment (pH 2 to 3.5) in vertebrates created by the secretion of hydrochloric acid (HCl) mainly functions to kill bacteria and to aid digestion by solubilizing food. The pH regulatory mechanisms in vertebrate stomachs are well understood. Until present, though, the gastric pH regulation in early deuterostome invertebrate larvae (such as echinopluteus and hemichordate tornaria) has been of little interest. A contrasting feature of early deuterostome larval stomachs is their highly alkaline gastric pH between 9.5 (echinopluteus) and 10 (tornaria) and well adapted digestive enzymes (proteases and phosphatases). As determined using pharmacological, electrophysiological and molecular tools, the cellular mechanisms responsible for gastric alkalization are mainly based on proton excretion from the stomach lumen via the gastric cells to the extracellular matrix. In Strongylocentrotus purpuratus , Na + /K + -ATPase (NKA) and H + -pump (VHA) are driving secondary active transporters such as the Na + /H + -exchanger (NHE) and the K + /H + -exchanger (KHE). Hemichordate Ptychodera flava larvae additonally employ bicarbonate secreting transporters such as Na + /HCO 3 - -cotransporter (NBC3) and anion exchanger (AE) to archive an even higher pH within the stomach lumen than echinopluteus larvae. Another interesting difference between both larvae is the use of H + /K + -ATPase (HKA, detected by the alkalization inhibition by the specific inhibitor omeprazol) in P. flava instead of VHA – and the presence of a protein in the stomach epithelium that is detectable by a human HKA antibody. In vertebrates, HKA has evolved to the driving force and the major contributor to achieving such a strong acidic environment within the stomach. Here we provide evidence that basic characteristics of vertebrate gastric pH regulation are already established in echinoid and hemichordate larval stages. Page 36 of 51 Poster Abstracts 71 The small GTPase Arf6 is essential for the development of the sea urchin larval gut Ahiakonu, Priscilla (University of Delaware); Stepicheva, Nadezda (University of Delaware); Dumas, Megan (University of Delaware); Song, Jia L. (University of Delaware) Arf6 is a small GTPase that acts as a molecular switch, cycling between the plasma membrane in its active GTP bound form and endosomal compartments in its inactive GDP bound form. While a rich knowledge exists in the cellular functions of Arf6, relatively little is known about its physiological role in development. This study examines the function of Arf6 in mediating actin remodeling and membrane trafficking in early development using the purple sea urchin as a model. We found that perturbation of Arf6 in the form of its knockdown, the constitutively activated GTPase defective Arf6 mutant Arf6 (Q67L), and the dominant active Arf6 mutant Arf6 (T27N), resulted in severe gastrulation defects. Arf6 knockdown led to a range of dose-dependent severity of developmental defects including a delay in development, vegetal cell attachment, and exogastrulation. We tested perturbation of Arf6 during gastrulation by examining its effect in the migration of endodermal cells. Results indicate that Arf6 perturbed embryos have wider gut size, blastopore and disorganized endodermal cells, leading to severe defects in larval gut structures and function. Overall our results demonstrate that Arf6 is essential for proper morphogenic movements of endodermal cells during gastrulation and the development and function of the larval gut. 72 1-MA signaling and Hox cluster in the crown-of-thorns Acanthaster planci starfish. Baughman, Ken (OIST); Satoh, Nori (OIST); Shoguchi, Eiichi (OIST) AUTHORS: Kenneth Baughman, Nori Satoh, and Eiichi Shoguchi Crown-of-Thorns starfish Acanthaster planci are corallivorous starfish, known for their consumption and devastation of hard corals in the Pacific and Indian Oceans 1 . Investigation of A. planci developmental biology may be useful for understanding how to mitigate the damage A. planci causes to coral reefs. First, we explored A. planci 1-methyl-adenine (1-MA) signaling in oocyte meiotic resumption 2,3 , and mapped 1-MA signaling using our own A. planci sequencing data. We confirmed that 1-MA signaling functions in A. planci by using a 1-MA protocol to synchronize and fertilize eggs. We observed that 1-MA resulted in egg ejection from female gonad tissue. Subsequent fertilization in the presence of 1-MA, but not in its absence, confirmed 1-MA function in A. planci. Based on the existing starfish literature and our own genome and transcriptome data, we mapped 1-MA oocyte resumption in a Systems Biology Graphical Notation (SBGN) diagram 4 . We find acceptable candidate genes for all components in our SBGN diagram. Second, using our sequencing data, we identified the Hox cluster in A. planci , and explored its phylogenetic consequences. Based on our Hox cluster data, we conclude that A. planci retains the overall organization of the Hox cluster found in hemichordates 5 , but retains the uniquely echinoderm organization of Hox13a-c. 1. Chesher, R. H. Destruction of Pacific corals by the sea star Acanthaster planci. Science 165, 280–283 (1969). 2. Kishimoto, T. A primer on meiotic resumption in starfish oocytes: The proposed signaling pathway triggered by maturation-inducing hormone. Mol. Reprod. Dev. 78, 704–707 (2011). 3. Kishimoto, T. & Kanatani, H. Cytoplasmic factor responsible for germinal vesicle breakdown and meiotic maturation in starfish oocyte. Nature 260, 321–322 (1976). 4. Le Novère, N. et al. The Systems Biology Graphical Notation. Nat. Biotechnol. 27, 735–741 (2009). 5. Freeman, R. et al. Identical Genomic Organization of Two Hemichordate Hox Clusters. Current Biology 22, 2053–2058 Page 37 of 51 (2012). 73 A transcriptomic strategy for identifying novel mediators of the sea urchin larval immune response Buckley, Katherine M. (University of Toronto, Sunnybrook Research Institute); Ho, Eric (University of Toronto, Sunnybrook Resear); Rast, Jonathan (University of Toronto, Sunnybrook Resear) The purple sea urchin genome encodes a complex repertoire of genes involved in the immune response, including large gene families of pathogen recognition receptors, as well as signaling and effector molecules. Recently available transcriptome data add a further dimension by identifying expressed sequences more accurately than gene model predictions. Genes involved in the immune response are difficult to identify in a non-targeted screen for two reasons: (1) Many immune genes are transcriptionally activated only in response to specific pathogens. (2) Genes involved in immunity tend to evolve quickly and are therefore difficult to identify computationally based on sequence similarity. To identify genes that mediate the sea urchin immune response, we have generated RNA-Seq data from larvae responding to a gut-associated bacterial challenge. Larvae were exposed to the marine bacteria Vibrio diazotrophicus and collected at several time points. These data were used to assess expression levels of known transcripts throughout the course of infection. Several transcriptionally regulated genes were identified that are known to have important homologs in vertebrate immunity (e.g., IL-1 receptor, members of the Socs family, Nf-?B and PU.1). Additionally, we assembled the reads to generate the immune activated transcriptome. From this assembly, we have identified 184 previously unknown transcripts that are strongly activated in response to bacteria. Among these are a divergent homolog of IL-17, which is a key regulator of vertebrate epithelial immunity, and a transcript that encodes a peptidoglycan binding domain. Novel immune effectors and signaling molecules have been difficult to identify outside of vertebrates and this approach offers a new strategy for their discovery. 74 Expression of the karyopherin-alpha family of nuclear transport proteins in Lytechinus variegatus. Byrum, Christine (College of Charleston); Smith, Jason (College of Charleston); Easterling, Marietta (College of Charleston); Bridges, M. Catherine (College of Charleston) Among eukaryotes, karyopherin-alpha (KAP-α) importins act in concert with the KAP-β protein KPNB1 to transfer transcription factors and other molecules across the nuclear pore complex. Although many importins are ubiquitously distributed, differential expression of the KAP-α family has been reported in several instances and two of these, KPNA1 and KPNA2, play important roles in neural differentiation of mouse embryonic stem cells (Yasuhara et al., 2007 and 2012). To investigate potential roles of these nuclear transport proteins in developmental processes, we identified three members of the KAP-α family and examined mRNA distribution of these importins in embryos of the sea urchin Lytechinus variegatus. We found that LvKPNA1/5/6 and LvKPNA3/4 were clearly expressed in cleaving embryos. By the blastula and gastrula stages, expression was most evident in the vegetal plate and archenteron, and by the prism/pluteus stages, faint staining was observed only in the oral surface and/or gut. A third karyopherin, LvKPNA2/7, exhibited a different expression pattern and was first observed in the vegetal plate and in patches of ectodermal cells within the mesenchyme blastula. By the gastrula stage, it was found in the archenteron and several additional patches of ectodermal cells. In prism/pluteus stage embryos, LvKPNA2/7 was restricted to the gut and was localized to subsets of cells along the ciliary band. Similar patterns have been reported for the neural marker Synaptotagmin and for another gene that influences neural differentiation in vertebrates, LvBrn1/2. Our study is an important first step towards better understanding roles of these nuclear transport proteins in embryogenesis. Page 38 of 51 75 Transcriptome Analysis of Late Gastrula Pigment Cells Calestani, Cristina (Valdosta State University); Barsi, Julius C (California Institute of Technology); Tu, Qian (California Institute of Technology); Ortiz, Antonio (Valdosta State University); Buckley, Kate M (University of Toronto); Stearnes, Ariel (Valdosta State University); Rast, Jonathan P (University of Toronto); Davidson, Eric H (California Institute of Technology) Pigment cells derive from the non-skeletogenic mesenchyme and migrate during early gastrula to the aboral ectoderm. Several evidences support a role of pigment cells in the immune-defense of the larva. In order to identify a comprehensive set of genes required for pigment cells development, with a particular focus on differentiation genes and their direct upstream regulators, we sequenced the transcriptome of a late gastrula pigment cell enriched sample (39-46 hours p.f.). The transcriptome sequence has also the potential to shed more light on pigment cells function. The RNA-seq approach involved the comparison of a pigment cell enriched with a pigment cell depleted sample. Pigment cells were labeled by injecting fertilized eggs with a BAC containing gfp driven by the pigment cell specific promoter of glial cell missing (gcm). Embryos were disaggregated and GFP positive cells were isolated by FACS. The GFP negative fraction was also collected. RNA and cDNA libraries were prepared from both the GFP positive and the GFP negative fractions. The cDNA libraries were then sequenced. An estimate of the pigment cell genes enrichment was provided by the ratio between the transcript number, expressed as fragments per kilobase of transcript per million mapped reads (FPKM), of the GFP positive over the GFP negative fraction. Genes known to be exclusively expressed in pigment cells, such as gcm, six1/2, sulfotransferase1, flavin monooxygenase1, 2 and 3 were very effectively enriched, within a range of 50 to 250 times. Genes enriched at least 10 times belong to a wide range of functional categories including: cell adhesion, defensome, GPCR Rhodopsins, immunity, metalloproteases, nervous system, cell signaling, and transcription factors. 76 mTOR regulation of polysomal recruitment at fertilization Chassé, Héloïse (équipe TCCD, UMR 8227, Station Biologique de Roscoff, CNRS/UPMC); Boulben, Sandrine (équipe TCCD, UMR 8227, Station Biologique de Roscoff, CNRS/UPMC); Cormier, Patrick (équipe TCCD, UMR 8227, Station Biologique de Roscoff, CNRS/UPMC); Morales, Julia (équipe TCCD, UMR 8227, Station Biologique de Roscoff, CNRS/UPMC) In sea urchin eggs, fertilization triggers a 10-fold increase in protein synthesis rates while initiating the synchronous divisions of early development. Sea urchin eggs contain the initiation factor eIF4E associated to its inhibitor 4EBP. Fertilization induces a rapid dissociation of eIF4E from 4EBP, eIF4E then associates to the scaffold protein eIF4G, initiating the formation of a functional initiation complex. These events are dependent upon the activation of the mTOR pathway. Incubation of sea urchin eggs with mTOR inhibitors such as rapamycin or PP242, inhibits protein synthesis and the onset of cell cycle. The translatome corresponds to the subset of maternal mRNAs actively engaged in translation, which are recruited into polysomal fractions. We have analysed the role of the mTOR pathway on the translatome at fertilization using PP242 inhibitor: we show that the polysomal recruitment of some mRNAs are sensitive to mTOR inhibition, while others mRNAs are still translated despite the inhibition of the mTOR pathway. Among the translated mRNAs strongly dependent on mTOR activity are the mRNAs encoding proteins involved in the cell cycle. Page 39 of 51 77 SLC and Notch2 regulate dorsal-ventral PMC positioning and skeletal patterning Chung, Oliver (Boston University); Piacentino, Michael (Boston University); Hewitt, Finnegan (Boston University); Patel, Vijeta (Boston University); Ferrell, Patrick (Boston University); Chaves, James (Boston University); Li, Christy (Boston University); Hameeduddin, Hajerah (Boston University); Poutska, Albert (Max Planck Institute); Bradham, Cynthia (Boston University) In the early stages of sea urchin embryonic development, primary mesenchyme cells (PMCs) secrete a calcium carbonate skeleton in response to patterning cues from the adjacent ectoderm. To determine the molecular mechanism underlying skeletal patterning, we performed an RNA-seq-based screen, which identified several ectodermal patterning gene candidates, including the sulfate transporter, SLC26a2 (SLC), and the receptor, Notch2. SLC loss-of-function (LOF) results in PMC localization to the dorsal hemisphere, and results in ventral and anterior skeletal patterning defects. SLC is expressed in the ventral and anterior ectoderm, suggesting that SLC functions to provide an attractive cue to PMCs. Interestingly, Notch2 LOF promotes the reciprocal phenotype, with ventral PMC localization and dorsal skeletal patterning defects. Like SLC, Notch2 is also expressed in the ventral ectoderm. These data suggest that SLC establishes a gradient in sPGs that provides an attractive cue, while Notch2 functions as a molecular switch to direct a subset of PMCs to the dorsal territory. To test this model, we performed combined SLC LOF and Notch2 LOF experiments. Loss of both SLC and Notch2 produces embryos that lack an organized PMC ring; instead, the PMCs adopt an arbitrary and disorganized pattern within the embryonic blastocoel. These embryos typically fail to form a skeleton, or produce only small, rudimentary skeletal elements. Co-injection of control MO with either SLC or Notch2 MO does not provoke a similar outcome, but instead such embryos resemble the corresponding single MO phenotypes. These results support the model in which SLC and Notch2 function together to direct primary PMC migration and skeletal patterning. 78 Mesoderm and Micromere Development in Eucidaris tribuloides Coots, Ashley D. (Rochester Institute of Technology); Covington, Rae Ann (Rochester Institute of Technology); Lung, Kara (Rochester Institute of Technology); Wood, Maureen (Rochester Institute of Technology); Sweet, Hyla (Rochester Institute of Technology) Sea urchins are grouped into cidaroids (pencil urchins) and euechinoids (including pin-cushion type urchins, sand dollars, and heart urchins), which diverged about 250 million years ago. These groups have differences in both adult and embryo morphology. In the field of sea urchin development, euechinoids have been commonly studied, however, cidaroid development has not. The purpose of this study is to further document the normal development of the cidaroid sea urchin, Eucidaris tribuloides. The development of skeletogenic cells and pigment cells was examined. Both cell types accumulate slowly during development to about sixteen cells per embryo; however, the number of cells is quite variable. Micromere development during cleavage was also examined. In euechinoid embryos, four micromeres form at the 16-cell stage and form large micromeres and small micromeres at the next cleavage. The large micromeres develop into the skeleton, while the small micromeres contribute to the coelomic pouches. The micromeres also send signals to surrounding cells to make pigment cells and other cell types. In cidaroid embryos, the number of micromeres is variable in that they may have 0, 1, 2, 3, or 4 micromeres in one embryo. The micromeres of the 16-cell stage embryo do divide to form both large micromeres and small micromeres. The number of micromeres contained in cidaroid embryos was found to be directly correlated with the number of skeletogenic cells in later stage embryos. In future studies, further research will be done on how the variability of micromeres affects cell signaling in the cidaroid embryo. 79 Expression of embryonic skeletal development genes during adult arm regeneration of the brittle star Amphiura filiformis Page 40 of 51 Czarkwiani, Anna (University College London); Dylus, David (University College London); Oliveri, Paola (University College London) The brittle star Amphiura filiformis is becoming an important model for regeneration research due to the emerging molecular toolbox which will complement already existing anatomical and ecological data. Its arms regenerate within a few weeks and re-pattern all the essential components such as skeleton, podia, radial water canal and the nerve. Initially after amputation and wound epithelialisation, a blastema (mass of undifferentiated cells) will be formed. Then during regeneration the small arm will elongate and begin to differentiate metameric units following the invasion of the radial water canal. Our lab has already shown that gene expression analysis like qPCR and whole-mount in situ hybridization (WMISH) can be successfully employed to study genes involved in brittle star arm regeneration. We use this novel model system to understand the relationship between regeneration and embryonic development using skeletogenesis as a proxy for studying cell specification, morphogenesis and molecular mechanisms. Using the WMISH technique we have characterized the expression pattern of approximately 20 important gene candidates from the embryonic skeletogenesis GRN during arm regeneration in A. filiformis. We found that most of the genes, which are expressed in the skeletal lineage of the brittle star embryo, are also localized to the skeletal domains in adult arm regeneration. During the early stage of regeneration those genes are expressed in a subepithelial layer of mesenchymal cells, which is where spicules first appear, and then become restricted to different skeletal elements during differentiation. Altogether this data shows that the regeneration molecular network underlying skeletogenesis might be similar to the embryo skeletogenic network, consistent with the idea of the co-option of the larval skeleton form the adult. 80 Polarity of small micromeres and its impact on localization of plasma membrane proteins Espinoza, Jose A. (University of California, San Diego); Campanale, Joseph (Scripps Institution of Oceanography); Gokirmak, Tufan (Scripps Institution of Oceanography); Hamdoun, Amro (Scripps Institution of Oceanography) Micromeres (Mics) undergo a reduction in ATP binding cassette (ABC) transporter activity, presumably related to their later epithelial-mesenchymal transition (EMT). Overexpression of constitutively active Cdc42 causes accumulation of the fluorescent ABC-transporter substrate calcein in the Mics to decrease from 1.6 to 1.07 fold (Mics/rest of the embryo). Calcein accumulation in the whole embryo remained unchanged, suggesting that ABC-transporter down-regulation is linked to cellular polarity. Next, we investigated cellular polarity in small micromeres (Smics) by expressing fluorescent pleckstrin homology domain (PH) of phospholipase C-delta, which binds to phosphatidylinositol 4,5-bisphosphate (PIP2). Smics accumulate nearly 80% more PH fluorescence on their basal surface than macromeres and mesomeres, indicating that cellular polarity in the Smics is altered. To examine relations between reduction in ABC-transporter activity and cellular polarity we co-expressed Sp -Vasa with a suite of ABC-transporters with robust apical and basolateral membrane polarity. We compared fluorescence of each transporter in Smics to controls in cleavage stage embryos. The apical transporters Sp -ABCB1a and Sp -ABCB4a, basolateral transporter Sp -ABCC1, and a neutral membrane marker were 15 to 50% less fluorescent on apical membranes. The apical transporter Sp -ABCG2a and the basolateral transporter Sp -ABCC1ß were 50 and 5% brighter, respectively. A 50-80% increase in fluorescence on the basal surface of Smics was measured for all transporters. Finally, we immunolocalized ABCB1a and Vasa protein at 48 HPF and found reduced ABCB1a localization on the apical surface of the Smics, suggesting that protein accumulation remains low and cellular polarity remains altered through migration. In the future, we plan to study interactions between polarity generating molecules in Smics, including cdc42 and phosphatidylinositol lipids, and ABC-transporter activity. Page 41 of 51 81 Ophioplocus esmarki Embryo and Larvae Swimming Behavior Freyn, Alec W. (Rochester Institute of Technology); Coots, Ashley (Rochester Institute of Technology); Sweet, Hyla (Rochester Institute of Technology) Brittle stars can be distinguished based on mode of reproduction as brooding or non-brooding species. Brooding brittle stars keep developing young inside bursal pouches until the juvenile stage, while non-brooders allow embryos and larvae to project into the water column, where ocean currents can transport them away from their parents. Brittle star larvae are further characterized as feeding or non-feeding. In feeding larvae, ciliary bands are used to obtain nutrients from the environment. Non-feeding larvae also have ciliary bands, but primarily use them for swimming. The main purpose of this study was to observe swimming patterns and calculate swimming speeds of the embryos and larvae of the brooding, non-feeding brittle star, Ophioplocus esmarki . We hypothesize that swimming behavior will differ in comparison to those displayed in non-brooding brittle star species. Embryos and larvae were collected from bursal pouches of adult brittle stars and their swimming behavior was observed up to eight days after fertilization. Unlike other non-feeding brittle stars, the embryos and larvae of this brooding species do not swim vertically into the water column. Swimming patterns consisted of four categories: circular, linear, hovering, and still, which are similar to what has been found in other non-feeding brittle stars. Swimming speeds were calculated by distance traveled (in mm) per second. It was found that there was a difference in swimming patterns over time; the embryos were more active early in development (swimming in circular and linear patterns) rather than later. There was also a statistically significant decrease in swimming speed over the six-day period. The changes observed in swimming behavior may correspond to changes in ciliary band, skeleton, or nervous system development. 82 Identifying and measuring voltage gradients in normal and perturbed embryos Hadyniak, Sarah E. (Boston University); Schatzberg, Daphne (Boston University); Lawton, Matthew L. (Boston University); Bishop, Jacob (Boston University); Beane, Wendy (Tufts University); Levin, Michael (Tufts University); Bradham, Cynthia (Boston University) Embryonic patterning requires the formation of endogenous bioelectrical gradients in several developmental model organisms. Voltage gradients also play a significant role in wound healing and regeneration. To identify changes in the relative voltage of the embryo, we used DiBAC 4 , a bis-barbituric acid oxonol dye that exhibits enhanced fluorescence in depolarized cells. In control embryos, DiBAC 4 fluorescence decreases steadily over time, indicating a relative hyperpolarization as development proceeds. This change is consistent with prior observations that differentiated cells are hyperpolarized compared to undifferentiated cells. In addition, the PMCs are relatively hyperpolarized compared to ectoderm in control embryos. The embryo also exhibits a gradient of electrical activity across the ectoderm. To determine the orientation of this gradient, we injected Nodal mRNA plus a fluorescent dye into one blastomere at the four cell stage, which biases the labeled blastomere to a ventral fate, then imaged the ventral label in conjunction with DiBAC 4 . The results show that control embryos possess a dorsal-ventral gradient of polarization within the ectoderm at hatched blastula stage. We performed a pharmacological screen for ion channel inhibitors, and identified SCH28080 as an inhibitor of skeletogenesis, and Concanamycin A (ConA) as an inhibitor of dorsal-ventral specification. DiBAC 4 analysis shows that SCH28080, an H + /K + antiport inhibitor, generally depolarizes the embryo, including the PMCs, although only the PMCs are developmentally affected by SCH28080. We will also report the effect of ConA, a V-ATPase inhibitor, on the DiBAC 4 distribution overall and within the ectoderm. Together these data establish the existence of voltage differences within the embryo, and Page 42 of 51 an important role for voltage differences within the PMCs and the ectoderm during normal development. 83 Semi-dry sea urchin experiment using preserved egg and sperm supply Kiyomoto, Masato (Ochanomizu University) The sea urchin is a wonderful material for observing animal development. Here, I propose a supply of preserved egg and sperm to realize a semi-dry sea urchin experiment, neither temperature-controlled aquarium nor a large volume of seawater needed. Sea urchin gametes were preserved for one month in some cases. Spawned eggs could be kept for the time with fertilizing ability in antibiotics seawater as Epel reported. It is also possible to lengthen the preserved time of dry sperm by the addition of antibiotics. Then, the provider, such as the marine laboratories, collects and prepares them, each laboratory and school just receive and do experiment. We had supported high schools from the two projects supported by CoREF (Consortium for Renovating Education of the Future), one was an easy and simple fertilization experiment without taking care of urchins for the beginners and the other was a larval culture in a small bottle by each student to observe the metamorphosis to juveniles. We started new projects, one is for the experimental courses in the universities (Supply of Marine Bioresource supported by the Ministry of Education, Culture, Sports, Science & Technology in Japan) and the other is for all elementary and secondary educations (as A Gift from the Sea supported by The Nippon Foundation). Both include gametes, embryos, larvae and juveniles to observe important stages. 84 Development of the nervous system in a sea cucumber, Apostichopus japonicus: shift from bilateral to pentaradial symmetry Kondo, Mariko (The Univ. of Tokyo); Nagai, Akiko (The Univ. of Tokyo); Kikuchi, Mani (The Univ. of Tokyo); Omori, Akihito (The Univ. of Tokyo); Akasaka, Koji (The Univ. of Tokyo) The five-fold radial, or pentaradial symmetry in the adult body plan is a well-known feature of echinoderms. In contrast, the larval body displays bilateral symmetry, and the body plan changes at the metamorphosis stage. In an attempt to investigate how pentaradial symmetry is formed, we are using a sea cucumber Apostichopus japonicus as our model. The sea cucumber does not produce an adult rudiment before metamorphosis, so the whole larval body is modified into the adult body. We are especially interested in the formation of neural tissues, since it is relatively easy to observe its development, and could possibly provide clues to how pentaradial symmetry is formed; during early development, a larval nervous system is generated, but a separate adult nervous system with five radial nerves appears at metamorphosis, which is probably one of the earliest examples of pentaradial symmetry. However, the mechanism how the nervous system or an overall pentaradial body is established is unknown. In addition, the developmental process of the sea cucumber is the same as sea lilies that are considered the most basal group in echinoderms, so this type of development may be the ancestral form of echinoderms. Therefore, the study on sea cucumber development may reveal the evolution of echinoderm body patterning. We have cloned several neural tissue marker genes including musashi (a marker for neurogenesis) and soxB1 (marker for stem cells or precursor cells in the central nervous system). Using these markers for expression analysis and anti-neural protein antibodies to perform immunohistochemistry, we are aiming to identify in which developmental stage and from where the adult nervous system arises. We expect that this work will provide insights to the molecular basis of the shift from bilateral to pentaradial symmetry. 85 Expression of germ cell markers in hemichordate Ptychodera flava: implication to the embryonic origin of PGCs in Ambulacraria Page 43 of 51 Lin, Ching-Yi (Institute of Cellular and Organismic Biology, Academia Sinica, Taiwan); Yu, Jr-Kai (Institute of Cellular and Organismic Biology, Academia Sinica, Taiwan); Su, Yi-Hsien (Institute of Cellular and Organismic Biology, Academia Sinica, Taiwan) Specification of primordial germ cells (PGCs) during development is an essential step for animals to produce next generation. Hemichordates are the sister group of echinoderms, which together are referred to as the Ambulacraria, that is closely related to chordates. According to previous studies, the PGCs of echinoderms develop from the coelom of larva except some sea urchin species, which develop PGCs from small micromere lineage. To understand how the common ancestor of Ambulacraria developed their PGCs, we study the expression of germline specific genes vasa and nanos in a hemichordate acorn worm Ptychodera flava . We found maternal vasa and nanos transcripts are expressed ubiquitously during the cleavage stages; subsequently vasa and nanos transcripts are co-localized to the dorsal-vegetal region of the archenteron after gastrulation. However, we found cytoplasmic vasa protein signals distributed near the vegetal pole of the unfertilized eggs and subsequently localized to the 8 vegetal blastomeres of the 32-cell stage embryo. After 64-cell stage, the distribution of vasa protein becomes peri-nuclear in the most vegetal-tier blastomeres. During gastrulation, the number of vasa-positive cells decreases, and these cells are localized to the dorsal-vegetal region of the archenteron, where the vasa and nanos are expressed zygotically. In tornaria larvae, these cells start to proliferate and later contribute to the developing adult coelom. Our results show the evolutionary consistency of PGCs marker genes expression patterns between echinoderms and hemichordate P. flava . However, the enrichment of vasa protein in the vegetal blastomeres appears to be much earlier in P. flava , suggesting there are some variations in vasa protein regulation between echinoderms and hemichordates during early embryogenesis. 86 BioTapestry: Interacting With the Network Directly in the Web Browser Longabaugh, William (Institute for Systems Biology); Paquette, Suzanne (Institute for Systems Biology); Leinonen, Kalle (Institute for Systems Biology) BioTapestry is a well-established tool for building, visualizing, and sharing models of gene regulatory networks (GRNs), with particular emphasis on the GRNs that drive development. The Java-based BioTapestry Viewer has been used to provide an interactive online version of the sea urchin endomesoderm network since 2003. Newer web-browser technologies such as HTML5 Canvas have made it possible to provide an interactive graphical network model directly in a web browser, and we have now created a version of the BioTapestry Viewer using these technologies. At the same time, this new software architecture continues to support the traditional BioTapestry Editor desktop application. 87 How pluteus arms were evolved? Morino, Yoshiaki (University of Tsukuba); Koga, Hiroyuki (University of Tsukuba); Wada, Hiroshi (University of Tsukuba) Some lineage of the echinoderms experienced an amazing evolution in their larval forms, namely acquisition of the pluteus arms. In sea urchin, interaction between epidermis and mesenchyme was shown to be essential for development of the pluteus arms. We also reported that a similar interaction is probably essential for the development of larval arms of ophiopluteus. Thus, the interaction between epidermis and mesenchyme is key issue for understanding the origin of pluteus larvae. Based on the common gene expression pattern of transcription factors between larval and adult skeletogenesis, it was suggested that evolution of larval skeleton was achieved in part by co-option of the specification program of adult skeletogenic cells were co-opted to embryonic stage. Here, we asked whether the interaction between epidermis and mesenchyme was also co-opted for the pluteus evolution. We Page 44 of 51 focused the development of direct-type developer sea urchin Peronella japonica . We examined expression pattern of alx1 , fgfA / fgfr2 , vegf / vegfr , pax2/5/8 , otp , pea3 , tetraspanin , wnt5 and P4 antigens in several developmental stages from gastrula to juvenile. Based on these dates, we discuss multiple steps required for the evolution of pluteus arm. 88 5-LOX is Required for Skeletal Patterning in Sea Urchin Embryos Murray, Ian S. (Boston University); Patel, Vijeta (Boston University); Li, Christy (Boston University); Yu, Annie (Boston University); Hameeduddin, Hajerah (Boston University); Hewitt, Finnegan (Boston University); Poustka, Albert (Max-Planck Institut für Molekulare Genetik); Bradham, Cynthia (Boston University) Skeletal patterning in sea urchins requires ectodermal instruction of the skeleton-secreting primary mesenchymal cells (PMCs). At late gastrula stage, the PMCs migrate to form a ring-and-cords pattern and begin to secrete the skeleton in the form of two triradiates. A comprehensive screen for ectodermal genes involved in the direction of PMC migration and positioning identified multiple skeletal patterning genes, including lipoxygenase (LvLOX). LOX is a leukotriene generator which indirectly impacts multiple signaling cascades. LvLOX was knocked down using an antisense morpholino-substituted oligonucleotide (MO), and skeletal patterning defects were systematically evaluated. The resulting embryos exhibit defects in triradiate orientation about the DV axis, defects in 2° skeletogenesis on the left side, and loss of midline elements. LOX exists in three primary isoforms in mammalian cells (5-LOX, 12-LOX, and 15-LOX), but the sea urchin ortholog cannot be definitively classified by Bayesian phylogeny. To better classify LvLOX, embryos were treated with three isoform-specific LOX inhibitors and a pan-LOX inhibitor, and the skeletal defects of the resulting perturbants were compared to that of LvLOX morphants. The embryos treated by the 5-LOX inhibitor MK886 exhibited similar rotational defects as well as loss of midline and left side elements, while the other isoform-specific drugs did not mimic the LvLOX MO. The pan LOX inhibitor produced more severe defects than either MK886 or LvLOX MO. Together these data demonstrate that leukotrienes are required for normal skeletal patterning, particularly in the midline and the left side, and for regulating triradiate orientation about the DV axis. 89 Histological and molecular biological analysis on the adult nervous system of the feather star Oxycomanthus japonicus Omori, Akihito (Misaki Marine Biological Station); Kurokawa, Daisuke (Misaki Marine Biological Station); Akasaka, Koji (Misaki Marine Biological Station) Adult nervous system of feather stars is subdivided into three systems; oral, deeper-oral, and aboral systems. Although previous studies show the shapes and possible functions of neural cells in each nervous system, studies about detailed whole-body cell distributions of these nervous systems is still limited. To confirm whole-body structures of these nervous systems, we examined immunolocalizations and gene expression patterns of some neural markers as well as observing detailed histological structures in a feather star Oxycomanthus japonicus . Histological stainings revealed a large ganglion of aboral nervous system with highly condensed cell bodies. Synaptotagmin immunoreactivity was detected in the ganglion, the nerve rings, and the radial nerves of all three nervous systems. We found that the expression of pan-neural marker gene Elav is not expressed in cells around articulations in the aboral brachial nerves, which indicates the existence of non-neural cells in the aboral nervous system. We also found that the expression of neural stem cell marker gene musashi is limited in the oral nervous system, suggesting the existence of immature neural cells in the oral nerves. Page 45 of 51 90 Circadian clock in the S. purpuratus larva: a diverged time-keeping mechanism that drives 24h rhythmicity? Petrone, Libero (UCL); Lerner, Avigdor (UCL); Oliveri, Paola (UCL) A circadian clock is a 24h time-keeping mechanism that synchronizes several biological processes with local environment. In metazoans the circadian system is driven by a regulatory network of so called "clock genes" interconnected in a transcriptional-translational feedback loops (TTO) that generates rhythmicity at mRNA and protein level. The TTO is entrained to environmental cues such as light. Evolution of circadian clocks provides an ideal system to understand evolution of regulatory networks. Sea urchin and its molecular tools can facilitate the comprehension of the evolution of the time-keeping mechanism in bilaterians. For this purpose we identified and analyzed the expression of orthologous clock genes in the sea urchin larvae. Genome survey identifies almost all clock genes known in protostomes and deuterostomes, with exception of period and p-like cry, indicating that the last common ancestor of all bilaterians had a complex clock toolkit. Furthermore, protein domain analysis shows evolution of proteins consistent with the presence/absence of members of the clock toolkit. Quantitative gene expression data reveal that the circadian clock begins to oscillate consistently in the free-living larva. Spu-vCry and Spu-tim show an opposite 24h oscillation in both light/dark (L/D) and free running (D/D) conditions; several other genes consistently show oscillation in L/D condition only; while, neither Spu-clock, nor Spu-bmal have rhythmic expression. Interestingly, in-situ hybridization of all sea urchin clock genes together with cell markers (e.g. serotonin) suggest the presence of light perceiving cells in the apical organ with a molecular mechanism similar to the protostome one. Importantly, our study highlights differences in the architecture and gene regulation of the sea urchin larval circadian clock compared to other metazoan clocks. 91 Appearance of Order Level Traits in the Triassic Echinoid Fossil Record Petsios, Elizabeth (University of Southern California); Thompson, Jeffery (University of Southern California); Bottjer, David (University of Southern California) Echinoid diversity in the Triassic (252-201 million years ago) is poorly understood, impeding detailed study of the evolutionary history of this group during this crucial time period following the appearance of the two modern subclasses of regular echinoids, cidaroids and euechinoids. Fossil representatives of early cidaroids and euechinoids are scarce, making interpretation of the evolutionary timing of appearance of crown group skeletal characters difficult. Diversification of these two groups appears to have occurred sometime during the Permian, arising from the family Archaeocidaridae, though fossil representatives from this time are limited. Stem group cidaroids of the family Miocidaridae are known from the Permian and Early Triassic, but it is not until the Late Triassic that the first definitive euechinoid is found. Known Triassic echinoid diversity tracks the occurrences of rare sites of exceptional echinoid fossil preservation (Lagerstatten), where whole or partially complete tests can be found. However, a large distribution of disaggregated echinoid material can be found throughout Triassic deposits, representing an untapped pool of potential diversity information. Though species level differentiation of specimens requires considerable preservation of articulated test material, ordinal level characters can be identified allowing for distinction between ancestral cidaroids and euechinoids and the identification of character states. Presented here is a survey of known Triassic echinoid diversity from the literature and the earliest known occurrences of ordinal level cidaroid and euechinoid traits. Future work will aim to uncover additional disarticulated fossil material from deposits that may record the occurrences of ordinal level characters from previously unknown time periods. Page 46 of 51 92 Concanamycin A perturbs dorsal-ventral specification in sea urchin embryos Reidy, Patrick (Boston University); Bishop, Jacob (Boston University); Schatzberg, Daphne (Boston University); Zushin, Peter (Boston University); Ross, Erik (Boston University); Carney, Tamara (Boston University); Bradham, Cynthia (Boston University) Bioelectrical gradients are important for normal development, wound healing, and regeneration. Bioelectrical changes are mediated by differential ion channel activity. In a pharmacological screen for ion channel inhibitors, we found Concanamycin A (ConA), a V-ATPase inhibitor, blocks dorsal-ventral specification in sea urchin embryos. ConA treatment results in morphological and skeletal radialization, and ConA-treated embryos exhibit an unrestricted ciliary band that occupies most of the ectoderm. Timecourse experiments demonstrate that embryos remain sensitive to ConA until after mesenchyme blastula stage, indicating that the effect of ConA is subsequent to the onset of Nodal and BMP2/4 expression. In situ hybridization experiments show that ConA-treated embryos express BMP2/4 mRNA, but do not express BMP2/4 targets, while Nodal and Nodal targets exhibit expanded expression profiles. Taken together, these data indicate that ConA treatment ventralizes embryos and prevents BMP2/4 target gene expression. 93 LvBMP5-8 is required for normal skeletal patterning but not dorsal-ventral specification in the sea urchin embryo Ramachandran, Janani (Boston University); Chung, Oliver (Boston University); Piacentino, Michael (Boston University); Reyna, Arlene (Boston University); Yu, Jia (Boston Uniersity); Hameeduddin, Hajerah (Boston University); Poustka, Albert (Max Planck Institute); Bradham, Cynthia (Boston University) Skeletal patterning in the sea urchin embryo is established by the communication between the pattern-dictating ectoderm and the skeletogenic primary mesenchyme cells (PMCs); however, the molecular basis for this process remains unknown. To identify skeletal patterning genes, we performed a three-way differential RNA-seq screen. One candidate gene identified by this screen is bone morphogenetic protein 5-8 (BMP5-8), a member of the TGF-β superfamily of signaling ligands. At late gastrula stage, LvBMP5/8 is expressed in the ventral ectoderm. We performed BMP5-8 loss-of-function (LOF) analysis by morpholino antisense oligonucleotide (MO) microinjection, and found that BMP5-8 morphant plutei display rotational and left-sided skeletal patterning defects. Dorsal-ventral (DV) axis specification and neural development are normal in BMP5-8 LOF embryos, indicating that LvBMP5-8 is not required for dorsal specification. Consistent with that conclusion, combined injections of LvBMP2/4 and LvBMP5-8 MO appear equivalent to LvBMP2/4 MO alone. Interestingly, BMP5-8 gain-of-function (GOF) results in radialization and loss of skeletal elements, similar to the effects of BMP2/4 GOF, except that BMP5-8 GOF embryos exhibit an apical bellcap. BMP5-8 GOF results in a complete loss of ventral Chordin expression and a radialization of dorsal Tbx2/3 expression, indicating that BMP5/8 is sufficient for dorsalization. Immunostaining in BMP5-8 GOF embryos show that the ciliary band is restricted to the apical plate, and serotonergic neurons are restricted to the apical bellcap. Synaptotagmin B-positive neurons extend processes radially around the ectoderm in a circumferential belt, and occasionally occupy the endoderm in BMP5-8 GOF embryos. Thus, LvBMP5-8 is required for left-right but not dorsal-ventral specification. 94 H+/K+ antiport activity is required for PMC differentiation and skeletogenesis Schatzberg, Daphne (Boston University); Lawton, Matthew (Boston University); Hadyniak, Sarah (Boston University); Bishop, Jacob (Boston University); Ross, Erik (Boston University); Carney, Tamara (Boston University); Beane, Wendy (Tufts University); Levin, Michael (Tufts University); Bradham, Cynthia (Boston University) Page 47 of 51 The bioelectrical signatures associated with regeneration, wound healing, development, and cancer are low amplitude changes in the polarization state of the cell, which persist over long durations. To identify ion channels required for bioelectrical changes during normal development of the sea urchin Lytechinus variegatus , a pharmacological screen was performed . We identified SCH28080, a competitive inhibitor of the H + /K + antiport ATPase, as a potent inhibitor of skeletogenesis in the developing larva. The ectoderm is correctly specified and differentiated in SCH28080-treated embryos, indicating that SCH28080 exerts a PMC-specific effect on skeletogenesis. A panel of fluorescent dyes was used to asses the ion distributions in PMCs within SCH28080-treated embryos. Compared to controls, the PMCs in SCH28080-treated embryos are relatively depolarized and exhibit a relatively high proton concentration at time points corresponding to PMC ingression and early migration, but return to levels comparable to controls during gastrulation . Sodium and chloride ion concentrations exhibit comparatively late changes in SCH28080-treated embryos that are likely compensatory. In control embryos, overexpression of the transcription factor Pmar is sufficient to convert all the cells in the embryo to PMCs. However, this effect is incomplete in SCH28080-treated embryos, indicating that SCH28080 impairs PMC differentiation. While PMC ingression is delayed but otherwise normal in SCH28080-treated embryos, the PMCs fail to undergo their final cell cleavage, to migrate to their typical positions, and to secrete a skeleton. SCH28080-treated embryos also exhibit defective PMC syncytium formation. These data indicate that activity of the H + /K + antiport ATPase is required for the set of changes that hallmark PMC differentiation following ingression. 95 Genome-wide analysis of the skeletogenic gene regulatory network of sea urchins Shashikant, Tanvi (Carnegie Mellon University); Rafiq, Kiran (Carnegie Mellon University); Ettensohn, Charles (Carnegie Mellon University) A central challenge of developmental and evolutionary biology is to understand the transformation of genetic information into morphology. Elucidating the connections between genes and anatomy will require model morphogenetic processes that are amenable to detailed analysis of cell/tissue behaviors and to systems-level approaches to gene regulation. The formation of the calcified endoskeleton of the sea urchin embryo is a valuable experimental system for developing such an integrated view of the genomic regulatory control of morphogenesis. A transcriptional gene regulatory network (GRN) that underlies the specification of skeletogenic cells (primary mesenchyme cells, or PMCs) has recently been elucidated. In this study, we carried out a genome-wide analysis of mRNAs encoded by effector genes in the network and uncovered transcriptional inputs into many of these genes. We used RNA-seq to identify >400 transcripts differentially expressed by PMCs during gastrulation, when these cells undergo a striking sequence of behaviors that drives skeletal morphogenesis. Our analysis expanded by almost an order of magnitude the number of known (and candidate) downstream effectors that directly mediate skeletal morphogenesis. We carried out genome-wide analysis of (1) functional targets of Ets1 and Alx1, two pivotal, early transcription factors in the PMC GRN, and (2) functional targets of MAPK signaling, a pathway that plays an essential role in PMC specification. These studies identified transcriptional inputs into >200 PMC effector genes. Our work establishes a framework for understanding the genomic regulatory control of a major morphogenetic process and has important implications for reconstructing the evolution of biomineralization in metazoans. 96 Expression of the ATP-Binding Cassette transporter Sp-ABCC5a in pigment cells is required for sea urchin gastrulation Shipp, Lauren E. (Scripps Institution of Oceanography); Hill, Rose (Scripps Institution of Oceanography); Moy, Gary (Scripps Institution of Oceanography); Gokirmak, Tufan (Scripps Institution of Oceanography); Hamdoun, Amro (Scripps Institution of Oceanography) Page 48 of 51 ATP-binding cassette (ABC) transporters are synthesized during development with roles in morphogenesis and protection from xenobiotics. In embryos of the sea urchin ( Strongylocentrotus purpuratus ), one such transporter is Sp -ABCC5a, whose expression in a subset of mesodermal cells is required for orientation and development of the endodermal hindgut. Transcripts of the ABCC5a gene are first detected at hatching (21 hpf) and are most abundant during the gastrula stage (42 hpf). Expression is limited to a subset of aboral non-skeletogenic mesenchyme (NSM) cells that become pigment cells. ABCC5a protein expression is first detected during early gastrulation (34 hpf) and peaks at the prism stage (50 hpf). ABCC5a expression is controlled by Delta-Notch signaling emanating from the skeletogenic mesenchyme, as treatment with DAPT at 3 hpf, but not 17 hpf, blocks its induction. Morpholino knockdown of ABCC5a (~210 kDa) does not appear to affect differentiation of pigment and blastocoelar cells, or production of echinochrome pigment, but instead results in abnormal archenteron formation. ABCC5a-morphants develop elongated archenterons and fused mouths, but following gastrulation, their hindguts protrude out from the blastopore to form a prolapse of the vegetal/posterior pole. This is observed in 90% of knockdown embryos by the late prism stage (~60 hpf). We hypothesize that efflux of signaling molecules, possibly cyclic nucleotides, from ABCC5a is required for complete invagination of the endoderm. In the absence of ABCC5a, hindgut-precursor cells remain on the vegetal pole of the embryo throughout gastrulation. This would suggest that pigment cell precursors help orient the endoderm cells during gut formation. We are currently addressing this hypothesis by further characterizing the ABCC5a-knockdown phenotype with live-imaging and cell-type labeling, and by probing the potential substrates of ABCC5a with efflux and migration assays. 97 microRNA-31 Regulates Skeletogenesis of the Sea Urchin Embryo Stepicheva, Nadezda (University of Delaware); Song, Jia (University of Delaware) The microRNAs (miRNAs) are small non-coding RNAs that regulate the translation and stability of target mRNAs. miRNAs play an important role in early developmental processes in many organisms. However, the molecular mechanism of miRNA-mediated regulation in the embryo is difficult to study. Here we examine miR-31, which is a highly evolutionary conserved miRNA that was shown to play a role in human cancer and bone formation, in early development using the sea urchin as a model. Our data indicate that loss-of-function of miR-31 in sea urchin embryos disrupts the function and localization of primary mesenchyme cells (PMCs), which are responsible for the formation of skeletal spicules. Using luciferase reporter constructs and site-directed mutagenesis, we identified miR-31 to directly regulate Pmar1 , Alx1 and Cyclophilin 1 ( Cyp1 ) within the skeletogenesis gene regulatory network. Blocking the specific regulation of miR-31 on Alx1 in the developing embryo with miRNA target protector morpholino antisense oligonucleotides (miRNA TP MASO) resulted in mislocalization of PMCs. These results indicate that miR-31 mediated regulation Alx1 gene is sufficient to disrupt proper skeletogenesis. This study contributes to an understanding of the novel regulatory role of miR-31 in the developing embryo and incorporates post-transcriptional regulation by miR-31 into the skeletogenesis GRN. 98 miR-124 Regulation of the Delta/ Notch signaling pathway Suarez, Santiago N. (University of Delaware); Song, Jia (University of Delaware) microRNAs (miRNAs) play a crucial role in cell differentiation. The brainspecific miR-124 is highly conserved in both invertebrates and vertebrates. We propose to use the purple sea urchin, Strongylocentrotus purpuratus, to examine the function of miR-124. The Delta/Notch is a conserved signaling pathway that regulates neural differentiation and it also activates GCM that is critical for the specification of mesodermally-derived pigment cells. We bioinformatically identified Delta, Notch and GCM to contain potential miR-124 binding sites. We hypothesize that miR-124 regulates the Delta/Notch Page 49 of 51 signaling pathway important for neurogenesis and mesodermal specification. We will examine loss-of-function and gain-of-function of miR-124 induced phenotypes and identify cell types regulated by miR-124 using various molecular markers. We observed a significantly larger midgut width in larval stage embryos in miR-124 knockdown embryos as compared to the control embryos. We will investigate the direct regulation of miR-124 on Delta, Notch, and GCM by cloning their 3’untranslated regions downstream of luciferase reporter constructs. This study elucidates the function of miR-124 by identifying its direct gene targets and contributes to our understanding of neural development and mesodermal specification. 99 Spatial Regulation of Gene Expression in the Skeletogenic Mesenchyme by Extrinsic Cues Sun, Zhongling (Carnegie Mellon University); Ettensohn, Charles (Carnegie Mellon University) Short-range signals from ectodermal territories regulate gene expression in the primary mesenchyme cells (PMCs) of the sea urchin embryo. Previous studies have shown asymmetric gene expression in PMCs after gastrulation despite the fact that PMCs are homogeneous when they are first specified. Recent studies have demonstrated the critical role of the VEGF and MAPK pathways in skeletogenesis. In this study, we use whole mount in situ hybridization to examine the expression patterns of 34 highly expressed PMC-specific/enriched mRNAs in Strongylocentrotus purpuratus embryos at the late gastrula, early prism and pluteus stages. The expression patterns of these 34 genes at each stage can be classified into 3-4 categories. Most of these mRNAs are expressed at higher levels at sites of rapid biomineral deposition. Furthermore, we use a VEGFR inhibitor, axitinib and a MAPK inhibitor, U0126 to show that the VEGF and MAPK pathways regulate gene expression in the PMCs at the tips of extending arms at the pluteus stage in a similar way, suggesting that the MAPK pathway acts downstream of VEGF. Both pathways are essential for maintaining high expression level in the PMCs at the tips of the anterolateral rods and the postoral rods. Gene expression in the PMCs at the tips of the body rods on the dorsal side of the embryo is not affected by axitinib or U0126 treatment. We propose that the coordinate, localized expression pattern of suites of effector genes reflects regulation via a common signaling pathway. Our results indicate that multiple signaling pathways are involved in regulating gene expression in the PMCs: VEGF/MAPK signaling on the ventral side and a separate, unidentified pathway on the dorsal side. In addition, we find that the expression of individual effector genes can be sequentially regulated by multiple signaling pathways during development. 100 A probabilistic modeling of the cell lineage highlights interindividual variability in Paracentrotus lividus early development villoutreix, paul (cnrs); Rizzi, Barbara (CNRS); Delile, Julien (CNRS); Duloquin, Louise (CNRS); Faure, Emmanuel (CNRS); Savy, Thierry (CNRS); Bourgine, Paul (CNRS); Peyriéras, Nadine (CNRS) Assessing the question of inter-individual specific differences and intra-individual cell differentiation at the single cell resolution is renewed by novel live imaging and reconstruction techniques. The sea urchin Paracentrotus lividus is used as a model to describe, measure and quantify it. We studied a cohort of five digitally reconstructed developing embryos from the 32 cells stage (4hpf) to hatching (around 500 cells, 10 hpf) which consist in the automatic reconstruction of the spatio-temporal cell lineage (3D+time) and the membrane segmentation of each cells from 2-photon microscopy processed by the BioEmergences platform. A first level of interindividual macroscopic variability is captured by a linear spatio-temporal rescaling. A finer level of description was defined by clustering cells by common cell type and cell cycle, allowing interindividual comparison. These groups were the basis for a probabilistic modeling reproducing the cellular proliferation, volume and surface macroscopic dynamics. This model show the independency of the life length characteristics between a cell and its mother, and the Page 50 of 51 decomposition of the cellular volume and surface into a stochastic and deterministic part. We produced a prototypical model able to predict the behavior of a normal blastula by merging the parameters value from each specimen of the cohort. The corresponding artificial cell lineage could be embedded in space with a biomechanical model. In addition to testing mechanical assumptions, this spatio-temporal prototype may be used as a spatial template for a gene expression atlas integrating individual cell variability. This work is one the first attempt to study the variability from the individual cell to the whole organism, thus contributing to the foundation of the emerging field of quantitative developmental biology. 101 The embryonic transcriptome for Lytechinus variegatus Zuch, Daniel (Boston University); Hogan, J.D. (Boston University); Keenan, Jessica (Boston University); Luo, Lingqi (Boston Univesity); Saji, Akhil (Boston University); Sundermeyer, Mary Ann (Boston Univesity); Piacentino, Michael (Boston Univesity); Schatzberg, Daphne (Boston Univesity); Azzizi, Elham (Boston Univesity); Zhang, Shile (Boston Univesity); Heilbut, Adrien (Boston Univesity); Poustka, Albert (Max Planck Institute, Berlin Germany); Bradham, Cynthia (Boston Univesity) The transcriptome for L. variegatus (Lv) was sequenced at 11 developmental timepoints using the Illumina platform, then assembled using SOAPdenovo trans and annotated by BLAST analysis against S. purpuratus (Sp) gene models and nr. The N50 for the assembled scaffolds was 638. The timepoints were normalized using quantile normalization, and the expression data were validated using qPCR analysis. We identified sequences that correspond with 18825 S. purpuratus gene models. We evaluated the stage of onset for known endomesodermal and ectodermal gene regulatory network genes, and find that the onsets for the majority of these genes in Lv agrees with predictions from both Sp and P. lividus, suggesting that the overall network logic is preserved among these species. K-means clustering identified numerous clusters within the overall transcriptome as well as among the developmentally expressed transcription factors, including clusters expressed primarily at single stages, and clusters with broader expression profiles. Principal component (PC) analysis identified the transition from early blastula to hatched blastula as the transition accounting for the most variation (PC1), followed by the transition from late gastrula to early pluteus (PC2), then the transitions from mesenchyme blastula through early, mid and late gastrula (PC3). Together, the first three PCs account for 71% of the variation in the expression data. Finally, we analyzed the vertebrate phylotypic period genes identified by Irie and Kuratani (Nature Communications 2011). Of the 109 genes, we identified 93, and found that the largest number are co-expressed at "late" pluteus stage (48 hours post-fertilization), suggesting that sea urchin plutei correspond most closely to the vertebrate pharyngula stage in terms of gene expression profiles. Page 51 of 51 Participants Priscilla Ahiakonu University of Delaware pkahiako@udel.edu KOJI AKASAKA University of Tokyo kojiaka@mmbs.s.u-tokyo.ac.jp Carmen Andrikou Stazione Zoologica 'Anton Dohrn' di Napoli carmen.andrikou@szn.it M. Ina Arnone Stazione Zoologica Anton Dohrn miarnone@szn.it Julius Barsi Caltech barsi@caltech.edu Ken Baughman OIST kbaughman@oist.jp Smadar Ben-Tabou de-Leon The University of Haifa, Israel sben-tab@univ.haifa.ac.il Andrea Bodnar Bermuda Institute of Ocean Sciences andrea.bodnar@bios.edu Cynthia Bradham Boston University cbradham@bu.edu Bruce Brandhorst Simon Fraser University brandhor@sfu.ca Katherine Buckley University of Toronto, Sunnybrook Research Institute kbuckley@sri.utoronto.ca David Burgess Boston College david.burgess@bc.edu Robert Burke University of Victoria rburke@uvic.ca Christine Byrum College of Charleston byrumc@cofc.edu Cristina Calestani Valdosta State University ccalestani@valdosta.edu R. Andrew Cameron California Institute of Technology acameron@caltech.edu Joseph Campanale Scripps Institution of Oceanography jpcampan@ucsd.edu Gregory Cary CMU gregorycary@gmail.com Vincenzo Cavalieri University of Palermo vincenzo.cavalieri@unipa.it Héloïse Chassé équipe TCCD, UMR 8227, Station Biologique de Roscoff, CNRS/UPMC hchasse@sb-roscoff.fr Alys Cheatle Jarvela Carnegie Mellon University acheatle@andrew.cmu.edu Oliver Chung Boston University chungman27@gmail.com Ashley Coots Rochester Institute of Technology adc5793@rit.edu Jenifer Croce CNRS jeni.croce@obs-vlfr.fr Miao Cui Caltech miaocui@caltech.edu Anna Czarkwiani University College London a.czarkwiani@ucl.ac.uk Eric Davidson California Institute of Technology davidson@caltech.edu David Dylus University College London david.dylus.10@ucl.ac.uk Allison Edgar Duke University ae75@duke.edu Susan Ernst Tufts University susan.ernst@tufts.edu Eric Erkenbrack Caltech erkenbra@caltech.edu Jose Espinoza University of California, San Diego j1espino@ucsd.edu Charles Ettensohn Dept. of Biol. Sci., Carnegie Mellon University ettensohn@andrew.cmu.edu Roberto Feuda Caltech rfeuda@caltech.edu Constantin Flytzanis University of Patras kostas@bcm.tmc.edu Adam Foote Carnegie Mellon University afoote@andrew.cmu.edu Tara Fresques Brown University, Wessel Lab tara_fresques@brown.edu Alec Freyn Rochester Institute of Technology awf1659@g.rit.edu Feng Gao California Institute of Technology gaofeng@caltech.edu Sarah Hadyniak Boston University hadyniak@bu.edu John Henson Dickinson College henson@dickinson.edu Veronica Hinman Carnegie Mellon U veronica@cmu.edu Eric Ingersoll Penn State Abington epi1@psu.edu Masato Kiyomoto Ochanomizu University kiyomoto.masato@ocha.ac.jp Mariko Kondo The Univ. of Tokyo konmari@mmbs.s.u-tokyo.ac.jp Oliver Krupke University of Victoria okrupke@uvic.ca Patrick Lemaire CRBM, UMR5237, CNRS/University Montpellier; Institut de Biologie Computationnelle, Montpellier, France patrick.lemaire@crbm.cnrs.fr Thierry Lepage CNRS tlepage@unice.fr Enhu Li Caltech enhuli@caltech.edu Ching-Yi Lin Academic Sinica sunbeank@gmail.com William Longabaugh Institute for Systems Biology wlongabaugh@systemsbiology.org Chris Lowe Stanford University clowe@stanford.edu Deirdre Lyons Duke University dcl.duke@gmail.com Megan Martik Duke University megan.martik@duke.edu David McClay Duke University dmcclay@duke.edu Alex McDougall CNRS dougall@obs-vlfr.fr Daniel Medeiros University of Colorado, Boulder daniel.medeiros@colorado.edu Artemis Michail University of Patras amichail@upatras.gr Dolores Molina Institut de Biologie Valrose (iBV), UMR7277, CNRS/UNSA loli.molina.jimenez@gmail.com Kathleen Moorhouse Boston College moorhous@bc.edu Julia Morales equipe TCCD, UMR8227 CNRS-UPMC, Station Biologique de Roscoff morales@sb-roscoff.fr Yoshiaki Morino University of Tsukuba yoshiaki.morino@gmail.com Robert Morris Wheaton College rmorris@wheatonma.edu Ian Murray Boston University ismurray@bu.edu Jongmin Nam Rutgers University-Camden jn322@camden.rutgers.edu Paola Oliveri University College London p.oliveri@ucl.ac.uk Akihito Omori Misaki Marine Biological Station omori@mmbs.s.u-tokyo.ac.jp Nathalie Oulhen Brown University nathalie_oulhen@brown.edu Margaret Peeler Susquehanna University mpeeler@susqu.edu Margherita Perillo Stazione Zoologica Anton Dohrn di Napoli, Naples, Italy margherita.perillo@szn.it Isabelle Peter Caltech ipeter@caltech.edu Libero Petrone UCL libero.petrone.11@ucl.ac.uk Elizabeth Petsios University of Southern California petsios@usc.edu Nadine Peyrieras CNRS nadine.peyrieras@inaf.cnrs-gif.fr Michael Piacentino Boston University mpiacent@bu.edu Dominic Poccia Amherst College dlpoccia@amherst.edu Janani Ramachandran Boston University janani93@bu.edu Andrew Ransick Caltech andyr@caltech.edu Ryan Range Mississippi State University range@biology.msstate.edu Jonathan Rast University of Toronto jrast@sri.utoronto.ca Patrick Reidy Boston University preidy@bu.edu Laura Romano Denison University romanol@denison.edu Daphne Schatzberg Boston University daph@bu.edu Stephan Schneider Iowa State University sqs@iastate.edu Catherine Schrankel University of Toronto c.schrankel@gmail.com Nick Schuh Sunnybrook Health Science Center nick.w.schuh@gmail.com Tanvi Shashikant Carnegie Mellon University tshashik@andrew.cmu.edu Lauren Shipp Scripps Institution of Oceanography lshipp@ucsd.edu Charles Shuster New Mexico State University cshuster@nmsu.edu Stephen Small New York University sjs1@nyu.edu L Courtney Smith George Washington University csmith@gwu.edu Peter Smith University of Southampton p.j.smith@soton.ac.uk Jia Song University of Delaware jsong@udel.edu Andriana Stamopoulou University of Patras andriana.stamopoulou@gmail.com Nadezda Stepicheva University of Delaware nstepich@udel.edu Jonathon Stone Rochester Institute of Technology jjs8236@rit.edu Meike Stumpp Academia Sinica (ICOB) meike.stumpp@gmail.com Yi-Hsien Su Institute of Cellular and Organismic Biology, Academia Sinica yhsu@gate.sinica.edu.tw Santiago Suarez University of Delaware santisua@udel.edu Zhongling Sun Carnegie Mellon University zhonglin@andrew.cmu.edu S. Zachary Swartz Brown University Molec Biology, Cell Biology & Biochemistry steven_swartz@brown.edu Hyla Sweet Rochester Institute of Technology hxssbi@rit.edu Jeffrey Thompson University of Southern California thompsjr@usc.edu Sarah Tulin MBL stulin@mbl.edu Jon Valencia Caltech jev@caltech.edu Zheng Wei NIH/NIDCR zhwei@mail.nih.gov Michael Whitaker Newcastle University michael.whitaker@ncl.ac.uk Jennifer Wygoda Duke University jaw61@duke.edu paul villoutreix cnrs paul.villoutreix@inaf.cnrs-gif.fr Gary Wessel Brown University rhet@brown.edu Athula Wikramanayake University of Miami athula@miami.edu Dongdong Xu Brown university xudong0580@163.com Shunsuke Yaguchi University of Tsukuba yag@kurofune.shimoda.tsukuba.ac.jp Mamiko Yajima Brown University mamiko_yajima@brown.edu Vanesa Zazueta-Novoa Brown University vanesa_zazueta-novoa@brown.edu Minyan Zheng Carnegie Mellon University minyanz@andrew.cmu.edu Leonard Zon HHMI/Boston Children's Hospital zon@enders.tch.harvard.edu Daniel Zuch Boston University dtzuch@bu.edu