共用巨集
advertisement
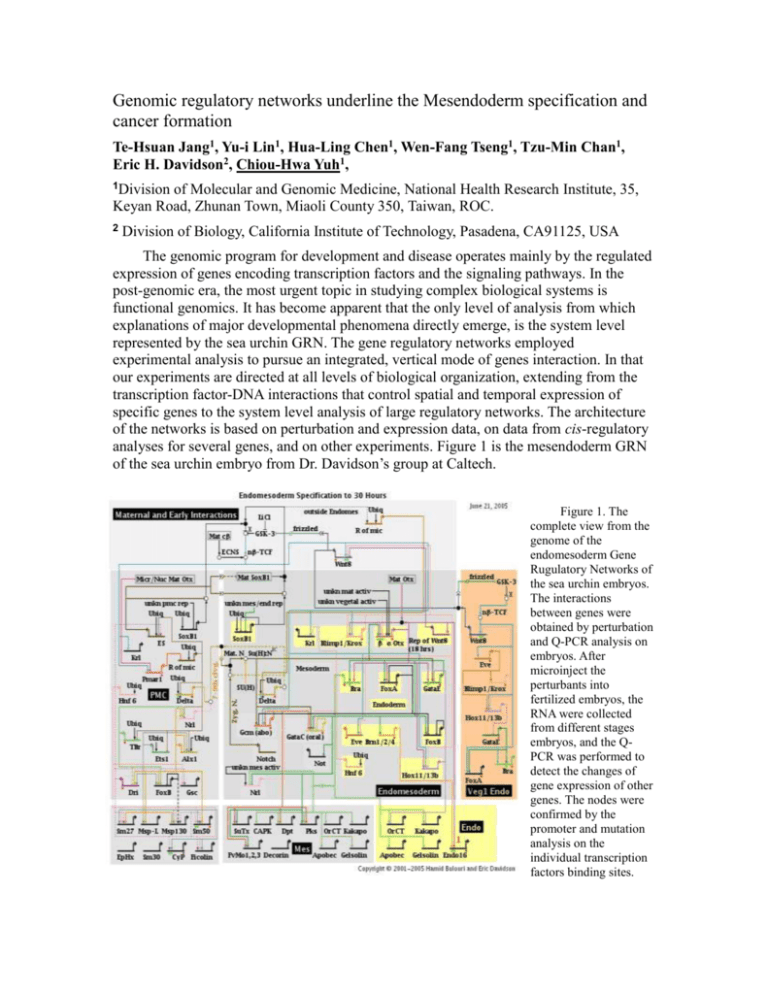
Genomic regulatory networks underline the Mesendoderm specification and cancer formation Te-Hsuan Jang1, Yu-i Lin1, Hua-Ling Chen1, Wen-Fang Tseng1, Tzu-Min Chan1, Eric H. Davidson2, Chiou-Hwa Yuh1, 1Division of Molecular and Genomic Medicine, National Health Research Institute, 35, Keyan Road, Zhunan Town, Miaoli County 350, Taiwan, ROC. 2 Division of Biology, California Institute of Technology, Pasadena, CA91125, USA The genomic program for development and disease operates mainly by the regulated expression of genes encoding transcription factors and the signaling pathways. In the post-genomic era, the most urgent topic in studying complex biological systems is functional genomics. It has become apparent that the only level of analysis from which explanations of major developmental phenomena directly emerge, is the system level represented by the sea urchin GRN. The gene regulatory networks employed experimental analysis to pursue an integrated, vertical mode of genes interaction. In that our experiments are directed at all levels of biological organization, extending from the transcription factor-DNA interactions that control spatial and temporal expression of specific genes to the system level analysis of large regulatory networks. The architecture of the networks is based on perturbation and expression data, on data from cis-regulatory analyses for several genes, and on other experiments. Figure 1 is the mesendoderm GRN of the sea urchin embryo from Dr. Davidson’s group at Caltech. Figure 1. The complete view from the genome of the endomesoderm Gene Rugulatory Networks of the sea urchin embryos. The interactions between genes were obtained by perturbation and Q-PCR analysis on embryos. After microinject the perturbants into fertilized embryos, the RNA were collected from different stages embryos, and the QPCR was performed to detect the changes of gene expression of other genes. The nodes were confirmed by the promoter and mutation analysis on the individual transcription factors binding sites. The overall goal of this project is to establish the Gene regulatory Networks underlying the mesendoderm development in zebrafish. The main reasons we use zebrafish as model system are: A, Zebrafish is vertebrate; the research of the GRN can apply to higher vertebrate organism, like human. B. Zebrafish embryos develop very fast; it can form a complete individual with well-defined organs within 48 hours after fertilization. C. We can easily knock down the genes’ function by injecting morpholino antisense oligonucleotide into the embryos. We also can make the transgenic animals very easily by microinjecting the cDNA clones of specific genes into the embryos. D. There are enormous efforts from all over the world using zebrafish for studying the gene regulation, cancer formation and developmental biology, but there are very few integrated studies regarding the gene regulation networks in zebrafish. According to literatures, the de-regulation of many transcription factors and signal transduction pathways are related to colorectal cancer formation. Those are the mutations of the components in PI3 kinase pathway, activation of Wnt pathway, changes at TGFpathway, and loss-of-function mutations in the JNK pathway. The lost function of FoxH1, smad2, smad4, CDX-2 and activation of SOX-9 and over expression of SNAIL gene product are also related to colorectal cancer formation. Interestingly, those transcription factors and signaling pathways are all in the mesendoderm networks we discovered from zebrafish model. We will combine the information we gathered from mesendoderm GRN and test their functions in the colorectal cancer formation. To understand the complex gene regulatory networks underlying the development and cancer formation, we need the help from computational modeling. In recent years, mathematical theories and computer simulations have started to become a useful tool in the Systems Biology study. We are going to collaborate with Professor Feng-Sheng Wang, hoping to establish the gene networks using reverse-engineering computational modeling method. References: 1. Yuh, C. H., Dorman E. R., Howard M. L. and Davidson, E. H. An otx cis-regulatory module: a key node in the sea urchin endomesoderm gene regulatory network. Dev Biol. 2004 May 15;269(2):536-51. 2. Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh C. H., Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan Z, Schilstra MJ, Clarke PJ, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A genomic regulatory network for development, Science, 295, 1669-1678, 2002. 3. Yuh, C. H., Bolouri, H. and Davidson, E. H. Genomic Cis-regulatory Logic: Experimental and Computational Analysis of a Sea Urchin Gene. Science, 279, 1896-1902, 1998. Useful link and software: http://labs.systemsbiology.net/bolouri/software/BioTapestry/#launch http://family.caltech.edu/tutorial/ http://sugp.caltech.edu/endomes/











