Sensing the Environment
advertisement

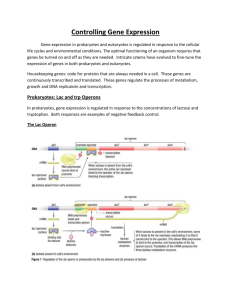
Regulating Gene Expression • Microbes respond to changing environment – Alter growth rate – Alter proteins produced • Must sense their environment – Receptors on cell surface • Must transmit information to chromosome • Alter gene expression – Change transcription rate – Change translation rate Sensing the Environment • Two‐component phosopho‐relay signal transduction – Receptor/Sensor Histidine‐kinase protein in plasma membrane • Binds to a signal cue • Activates itself via phosphorylation p p y – Cytoplasmic response regulator Takes phosphate from sensor Binds chromosome Alters transcription rate of multiple genes 1 Operonic regulation • Coding vs regulatory sequences • Regulatory sequences: promoters, operator and activator sequences • Regulatory proteins: repressors, activators – Repressors bind operator sequences, block transcription • Induction vs Derepression – Activator proteins bind sequences near by promoters, facilitate RNA Pol binding, upregulate transcription The E. coli The E. coli lac Operon • Lactose (milk sugar) is used for food – Cannot pass through plasma membrane • Lactose permease allows entry • PMF used to bring lactose inside cell – Must be converted to glucose to be digested β-galactosidase β galactosidase converts lactose to glucose and galactose People also make βgalactosidase If not, person is lactoseintolerant The E. coli The E. coli lac Operon • The lacZ gene encodes β‐galactosidase • The lacY gene encodes lactose permease – Need both proteins to digest lactose • Operon – Multiple genes transcribed from one promoter Multiple genes transcribed from one promoter 2 The E. coli The E. coli lac Operon • Repressor protein LacI blocks transcription – Repressor binds to operator – Blocks σ factor from binding promoter gp • Repressor responds to presence of lactose – Binds inducer (allolactose) or DNA, not both – Add lactose → repressor falls off operator Allolactose cause operon induction Activation of the lac Activation of the lac Operon by by cAMP cAMP‐‐CRP • Maximum expression requires cAMP and cAMP receptor protein (CRP) ‐ The cAMP‐CRP complex binds to the promoter at ‐60 bp ‐ Interacts with RNA pol, increase rate of transcription initiation • CRP acts as activator only when bound to cAMP Catabolite Repression • Glucose is present, lac operon is OFF (no transcription) • Two mechanisms involved 1. High glucose → low cAMP levels → CRP inactive – Can’t bind operon → low level of lac transcription • • • • • PTS dependent glucose uptake P-transfer from IIA-P to IIB for Glucose uptake IIA becomes available IIA: inhibitor of AC cAMP level reduced 3 Catabolite Repression 2. Inducer exclusion: – High glucose → high IIA – IIA inhibits LacY permease – Reducing intracellular lactose • Importance of catabolite repression – Sequential sugar catabolism – diauxic growth Animation: The E. coli lac Operon Arabinose operon • Regulation by dual role regulatory protein AraC • “AraC” acts as repressor to block transcription (no arabinose) • Acts also as activator when bound to “arabinose” (the inducer) – Operators O1, O2 and araI control AraC and AraBAD proteins expression 4 Ara Operon Controls • Senario I: No arabinose present – “AraC” forms long dimeric conformation, blocks transcription (binding O2, araI1) • Senario II: arabinose added – changes AraC dimeric conformation • acts as activator • Stimulates binding of RNA polymerase + arabinose Trp operon operon: Repression and Attenuation : Repression and Attenuation • trp operon – Cell must make the amino acid tryptophan • Trp operon codes and regulates biosythetic enzymes • When tryptophan is plentiful, cell stops synthesis • Regulation by two mechanisms 1. Repression: Trp repressor must bind tryptophan to bind DNA • Opposite of lac repressor Repressor + Tryptophan Transcription repressed Transcriptional Attenuation of the trp Operon 2. Attenuation: a regulatory mechanism in which translation of a leader peptide affects transcription of a downstream structural gene The attenuator region of the trp operon has 2 trp codons and is capable of forming stem-loop structures. 5 Transcriptional Attenuation Mechanism of the trp Operon High tryptophan Low tryptophan Animation: Transcriptional Attenuation Animation: Sigma Factor Regulation • • • • • σ factors regulate transcription of all genes Recognize promoter sequences differentially Specific to a subset of genes Direct RNA Pol to start transcription Alternative σ factors used for global transcriptional regulation of related genes 6 How are sigma factors regulated? • Temperature‐sensitive mRNA 2°‐structure – at 42°C σ70 degraded rapidly – Allows translation of σ32 (σΗ) only at high temperature • Synthesis of proteins that inhibit σ factors – Anti‐σ factors block σ activity until needed – Anti‐anti‐σ factors respond to environment Small Regulatory RNAs • found in bacterial intergenic regions • Regulate transcription or interfere with translation • antisense nature of sRNA allows its binding to mRNA ‐ ex. pairing of RNAIII with 5’‐end of mRNA prevents ribosome assembly, ie. halting translation – mRNA degradation RNA d d ti by dsRNA specific Read more: RNAIII represses virulence factors enzyme RNaseIII – RNAIII a multi‐locus global regulator of several vir. factors • Protein A (spa) • Coaggulase (Sa1000) • CRISPR system: Clustered Regularly Interspersed Short Palindromic Repeats • CRISPR arrays – Repeats: tandem array palindromes (4 to >100), 28‐40 bases, form stem‐loop in transcripts, 3’ terminus GAA(AC/G) – Spacers: 25‐40 bp, identical to phage or plasmid and some chromosomal seq., mostly sense, some antisense • Leader: ~550 bp, immediately 5’ to array , AT rich, promoter • CAS: clustered gene families, only present in genomes containing CRISPRs, code for RNA or DNA active proteins, possibly involved in processing arrays and psiRNA 1. CRISPERS: Nature Reviews (Microbiology), 6:181, 2008 2. Evolution of CRISPRS CAS prteins , Nature Reviews(Microbiology), 9:467, 2011 7 CRISPR: a global response to foreign DNA or RNA • specificity sensitive to single nucleotide difference • maintains memory • a primitive nucleic acid based adaptive immunity? Quorum Sensing • Cells work together coordinately at high cell density – V. fischeri becomes bioluminescent – Many bacteria form biofilms • Discovered in Vibrio fischeri, a bioluminescent bacterium that colonizes the light organ of the Hawaiian squid Quorum Sensing: Bioluminescence induction • Induction of a quorum‐sensing genes need accumulation of a secreted small molecule called an autoinducer – Homoserine lactone for V. fishcheri • At At a certain extracellular concentration, the secreted a certain extracellular concentration the secreted autoinducer reenters cells ‐ binds to a regulatory molecule, which in the case of Vibrio fischeri is LuxR ‐ The LuxR‐autoinducer complex then activates transcription of the luciferase target genes that confer bioluminescence 8 Quorum Sensing: Bioluminescence induction Animation: Quorum Sensing Animation: Summary ● Regulatory proteins help the cell sense and react to changes in its internal environment. ● Two‐component signal transduction systems help the cell sense its external environment. The lacZYA The lacZYA operon is regulated as follows: operon is regulated as follows: ‐ Operon is off when LacI binds to the operator. ‐ Operon is on when allolactose binds to LacI; cAMP‐ CRP are bound to the promoter (and there is no glucose around). ● The tryptophan operon is regulated by repression and attenuation (premature transcript termination). ● 9 Summary ● Sigma factors are controlled by alternate transcription and translation, proteolysis, and anti‐sigma factors ● Small regulatory RNAs can bind to mRNA and inhibit translation and cause mRNA degrade. CRISPR is a global reaction against invading foreign RNA or DNA reaction against invading foreign RNA or DNA Bacterial genes are regulated by a hierarchy of regulators that form integrated gene circuits. ‐ Example: Chemotaxis ● In quorum sensing, bacteria can communicate with each other at high cell densities via autoinducers ● 10