lab
advertisement

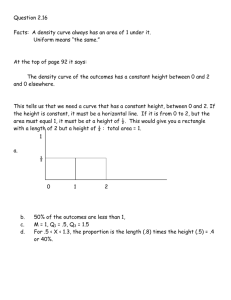
BE/EE189DesignandConstructionofBiodevices Project2:“On-chip”Real-timePolymeraseChainReaction(RT-PCR) Week3–OpticalDetectionandSystemTest Systemreadyforin-classtestingon:1Mar2016 Reportdue:10Mar2016 Instructions 1. Tobecompletedingroupsof2-3. 2. Yourthermocyclingshouldbeworkingbynow. 3. Handle all optics very carefully. Do NOT touch the surfaces of the lenses, filters, objectivelensesandcamerasensors.Whenindoubt,plsask. 4. Pleasefeelfreetoaskfordemonstrationsofhowtheopticsshouldbemounted.When indoubt(andifthere’snoonetoconsult),visitwww.thorlabs.comforsomeideas.We got all our opto-mechanic components there. It should get more intuitive as you start playingaroundwiththecomponents. 5. Onlyonelabreportisrequiredofeachgroup. 6. YourlabreportshoulddocumentclearlybutconciselyhowyourdeviceandVIwork.The labreportwillbegradedpartiallyonfunctionalityofthesystem.However,ifyourdevice doesn’t function perfectly, do not fret. A large emphasis will placed on understanding theshortcomingsandpointsoffailure(ifany)ofyoursystem. Opticaldetection Epifluorescencedetectionwillbeused.Theopticaldetectionsystem(essentiallyahomemade microscope) consists of an LED light source (see spec sheet attached), a filter set (excitation, dichroicandemission),anobjectivelens(infiniteconjugate,specsheetattached),atubelens andacamera. Figure1.Suggestedsetup. Q1.Whatisthefocallengthoftheplano-convexlens(50mm,75mmor100mm)?Howdidyou finditout(withoutreferringtoitspartnumber)? Q2.Howwouldyouorienttheplano-convexlens?Shouldtheflatsurfacefacetheobjectiveor thecamera?Hint:Oneorientationworksbetteratfocusingacollimatedlightintoapoint. Q3.Whatisthemagnificationofthemicroscopesysteminthesetup?Howdidyoufindout? ImportationConsiderations– 1. Sincefluorophorebleachingcanoccur,itisprudenttoturnontheLEDonlyduringsignal acquisition. 2. Atwhichpartofthecycleshouldthesignalacquisitionbedone? 3. How would you determine the background signal and eliminate/consider that in your data? 4. Since background light contributes to noise, how would you maximize the signal-tonoiseratio? Your LabView program should be able to acquire and plot a replication curve. A replication curveisrequired.Ameltcurveanalysisisnotrequired. Systemtest Atubeofcompletereactionmix(containingtemplateDNA,DNAprimers,polymerase,dNTPs, SYBRetc)willbegiventoyouatthebeginningofclass.We’retotestforthepresenceofaGMO gene.Here’sanexcerptfromtheBioRadGMOinvestigatorkitwe’reusing,justsowehavean ideaofwhatwe’retryingtoamplify: “TheGMOInvestigatorkitisdesignedtotestforthepresenceoftwodifferentGMO-associated DNA sequences: the 35S promoter of the cauliflower mosaic virus, and the terminator of the nopaline synthase gene of Agrobacterium tumefaciens. These DNA sequences are present in mostoftheGMcropsthatareapprovedfordistributionworldwide.” Testthefunctionalityofyoursystemduringtheclass. Atypicalamplificationplotlookslikethegraphinfigure2. Figure2.Typicalamplificationplot. Q4.Whywouldthecurvelooklikethis? Q5. Did you get a typical amplification curve? What are the sources of noise and design inadequacies?Howcanthesystembeimproved? PostlabQuestions Post-labQn1:Duetothetimeandresourcelimitations,weareonlyabletodoaveryrough simulationofwhatan“on-chip”RT-PCRdeviceshouldbeableto.(Weprobablyshouldn’teven callitanon-chipsystem.)Whatarethepotentialadvantagesofatrueon-chipplatform?Think alongthelinesofsamplepreparation,reagentmixing,reagentvolume,portability,etc… Post-labQn2:Withoutconsultingcurrentliterature,designafullyon-chipPCRdevice.(A conciseblockdiagramofdesignfeatureswilldo.Acompleteblueprintisnotrequired.)Thispart oftheexerciseisnotgradedbasedonfeasibility(butitcannotbecompletelyoutofthisworld). Justbeimaginativeandhavefun!Startfromanintacttissuesample.Theendpointinmind wouldbethedeterminationoftheabsenceorpresenceofaspecificDNAsequence. Post-labQn3:Readsomerecentpapersonon-chipPCR.Whichpartofyourimaginarydesign hasalreadybeentackled?(Citethepapersyoureferredto.)Whataretheremaining engineeringchallenges?(Ifthereisn’tanyremaining,thinkaboutanydesignopportunities. Includeassumptionsmade(e.g.availabilityoftechnologiesand/orprocesses).