Tom Walz Department of Cell Biology Harvard Medical School
advertisement

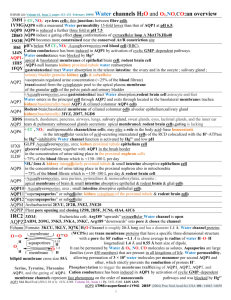
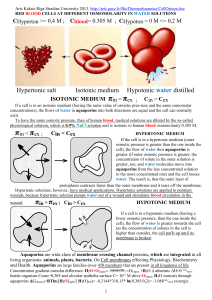
Tom Walz Department of Cell Biology Harvard Medical School Electron Crystallography Workshop UC Davis August 2006 The aquaporin family of water pores The occular lens Lens has to be transparent Lens must be able to accommodate Capsule Degrading nuclei Cortex Core Lens Lens fiber cells Cornea Conjuctiva Epithelium AQP0 forms thin junctions in vivo 0.2 µm Adapted from: Paul & Goodenough (1983) J. Cell Biol. 96: 625-632 Adapted from: Zampighi et al. (1982) J. Cell Biol. 93: 175-189 Purification of AQP0 from the lens cortex core Cortex 97 67 Core 45 31 full-length cleaved 21 Solubilization in 1% DM Anion exchange (MonoQ) Gel filtration (S12) 14 2D crystals of cortical AQP0 200 nm AQP0 2D crystal in negative stain CTF plot of glucoseembedded 2D crystal Projection map at 4 Å resolution AQP0 AQP1 How to get double-layered 2D crystals of AQP0 ? AQP0 in the lens cortex is full-length Some AQP0 in the lens core is C-terminaly cleaved AND AND Cortical AQP0 forms single-layered crystals AQP0 membrane junctions are more frequent in the core Cleavage of AQP0 may induce the formation of membrane junctions Chymotrypsin treatment of reconstituted AQP0 reconstituted cortical AQP0 chymotrypsin treatment membranes after cleavage Cleavage of AQP0 does indeed induce the formation of membrane junctions Purification of AQP0 from the lens cortex core Cortex 97 67 Core 45 31 full-length cleaved 21 Solubilization in 1% DM Anion exchange (MonoQ) Gel filtration (S12) 14 2D crystals of core AQP0 1 µm AQP0 2D crystal in negative stain CTF plot of glucoseembedded 2D crystal Projection maps of AQP0 2D crystals p422 p4 Double-layered crystal Single-layered crystal Electron diffraction of double-layered MIP 2D crystals 0 degree 70 degree AQP0-mediated membrane junction The water pores in AQP1 and AQP0 AQP1 AQP0 Electron diffraction at liquid He temperature The 1.9 Å density map The AQP0 water pore at 1.9 Å resolution CS-I Arg 187 Phe 48 NPAE Asn 164 NPAB Asn 68 His 66 CS-II Tyr 149 Phe 75 The water pores in the 1.9 Å EM and 2.2 Å X-ray structure 1.9 Å EM structure 2.2 Å X-ray structure Conformational switch of loop A upon proteolytic cleavage 2.2 Å X-ray structure 1.9 Å EM structure Conformational switch of loop A upon proteolytic cleavage 2.2 Å X-ray structure Conformational switch of loop A upon proteolytic cleavage 2.2 Å X-ray structure 1.9 Å EM structure Conformational switch of loop A upon proteolytic cleavage 2.2 Å X-ray structure 1.9 Å EM structure Constriction site I - the ar/R site AQP0 Ala 181 His 172 Arg 187 Phe 48 AQP1 Cys 191 His 182 Arg 197 Phe 58 Molecular dynamics using AQPZ UP Arg 189 DOWN Arg 189 Adapted from: Wang et al. (2005) Structure 13: 1107-1118 Arg 187 and Met 176 in AQP0 2.2 Å X-ray structure 1.9 Å EM structure Arg 187 and Met 176 in AQP0 2.2 Å X-ray structure 1.9 Å EM structure The packing of AQP0 in the 2D crystals The packing of AQP0 in the 2D crystals The packing of AQP0 in the 2D crystals The lipids surrounding an AQP0 monomer Protein-lipid interactions PC 1 PC 5 PC 6 Harvard Medical School Tamir Gonen Yifan Cheng Stephen Harrison Piotr Sliz University of Auckland Joerg Kistler Kyoto University Yoshinori Fujiyoshi Yoko Hiroaki