High Throughput Homogenous Bioluminescent Assays
advertisement

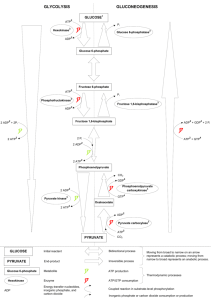
High Throughput Homogenous Bioluminescent Assays For Monitoring The Concentrations of AMP, ADP and ATP 1,2 Goueli , 1 Hsiao , 1 Zegzouti Said A. Kevin and Hicham Cell Signaling, Research and Development, Promega Corp.1 and University of Wisconsin School of medicine and Public Health2, Madison, WI 1. Abstract 4. Kinase Activity By Monitoring ADP Produced (ADP-Glo™ Assay) ADP conversion curves at different ADP/ATP concentrations Adenine nucleotides are major determinants of the energy status of the cell and thus any modulation of their cellular concentration has significant consequences to cellular metabolism, cellular growth and cell death. ATP generating enzymes are usually involved in anabolic processes while ATP consuming enzymes are involved in catabolite processes. We have developed biochemical assays that monitor the concentrations of ATP, ADP and AMP in a biochemical reaction. For instance ATP depletion dependent assays and ADP generating assays monitor the activity of kinases and ATPases. These universal assays can measure the activity of various enzymes with no modification of the native substrate and the ability to use diverse substrates such as proteins, peptides, lipids, sugars, etc. The AMP generating reactions such as ubiquitin ligases, aa aminoacyl t-RNA synthetases, DNA ligases, cAMP-dependent phosphodiesterases, etc. can be also monitored with high sensitivity and reproducibility. We will show data using these various technologies in monitoring the various adenine nucleotide concentrations in biochemical reactions. 10. Z’ Values, %CV, and S/B for the 16 Plates (AMP-Glo™ Assay) 7. Quantification of AMP in E3 Ub. Ligase Reaction 05.17 E3 Titration with 0.04µg Ubiquitin-WT with 100µM ATP Using AMP-Glo™ Ubiquitin Ligase (2µM, 1Hr Rxn) Study with 100µM ATP Using AMP-Glo™ 2.0E+06 R² = 0.9973 1.5E+06 RLU DRLU 6.0E+04 4.0E+04 1.0E+06 2.0E+04 5.0E+05 0.0E+00 0.0E+00 0 100 200 300 400 1.5µM AMP Rxn Buffer Average StDev %CV Average StDev %CV S/B ratio Z' Pl.1 378,510 15,181 4.0 594 617 103.7 637 0.875 Pl.2 369,828 17,942 4.9 821 757 92.3 451 0.848 Pl.3 363,068 18,639 5.1 783 756 96.6 464 0.839 Pl.4 367,053 16,428 4.5 832 816 98.0 441 0.859 Pl.5 361,599 19,385 5.4 855 811 94.9 423 0.832 Pl.6 356,236 14,775 4.1 720 723 100.3 495 0.869 Pl.7 347,899 16,438 4.7 704 701 99.6 494 0.852 Pl.8 340,925 16,976 5.0 564 554 98.2 604 0.845 Pl.9 347,537 16,596 4.8 762 761 99.8 456 0.850 Pl.10 344,758 15,765 4.6 884 829 93.8 390 0.855 Pl.11 343,265 17,694 5.2 570 593 104.0 602 0.840 Pl.12 335,801 18,563 5.5 684 674 98.6 491 0.828 Pl.13 337,908 17,757 5.3 720 690 95.9 470 0.836 Pl.14 334,863 16,817 5.0 691 685 99.2 484 0.843 Pl.15 334,951 17,507 5.2 649 667 102.8 516 0.837 Pl.16 331,386 15,516 4.7 638 628 98.4 519 0.854 68,184 78 2,834 1576 704 99 496 0.848 500 E3 (nM) ADP-Glo™ measures ADP produced in a kinase Reaction Sum= Average 2. Quantification of Cellular ATP (Promega Kinase-Glo®) "Negative" Ctrl. AMP-Glo assay 8.0E+04 2.5E+06 "Positive" Controls 5. Quantification of AMP in Biochemical Reaction (DNA Ligase – T4, ATP substrate) 349,724 8. Quantification of AMP in cAMP-PDE Reaction (cAMP-Phosphodiesterase PDE4B2) 16,999 4.9 T4 DNA Ligase Titration with 0.5µg DNA (pBR322)/Rxn with 100µM ATP Using AMP-Glo™ y = 5E+06x + 410472 R² = 0.9986 (in Buffer 1) Using K-R Buffer 120 3.0E+05 2.0E+07 y = 3E+06x + 376284 R² = 0.9959 (in BUffer 2) 1.0E+07 100 2.5E+05 R² = 0.9984 Activity % RLU AMP-Glo™ vs. LOPAC Using Buffers 3.0E+07 0.0E+00 2 4 6 8 2.0E+05 10 DRLU 0 ATP, µM ATP Level in HEK293 & CHO Using Kinase-Glo® 7.00 10K 5K 1.5E+05 80 60 LOPAC compounds +/- 3 St.Dev. +/- 25% 2.5K 1.0E+05 40 6.00 5.00 mM 717 11. LOPAC Library Screen Results (AMP-GloTM Assay) ATP Standard Titration Using Kinase-Glo® with Different Buffers 4.0E+07 Sum= 5.0E+04 20 4.00 0.0E+00 3.00 0 2 4 6 8 10 12 0 T4 Ligase, U/Rxn 2.00 0 200 400 600 800 1000 1200 Compound serial # 1.00 0.00 CHO HEK293 3. Kinase Activity using ATP Depletion (Promega Kinase-Glo®) 6. Quantification of AMP in Biochemical Reaction (DNA Ligase – E. coli, NAD substrate) 9. Quantification of Cellular ADP & ATP (CHO & HEK293) ATP & ADP Level in HEK293 & CHO Using AMP-Glo™ E. coli DNA Ligase Titration with 10µM Oligos & 10µM NAD Using AMP-Glo™ 7.00 200,000 120,000 R² = 0.9919 NADP + Oligos + Ligase (20min Rxn) RLU 140,000 80,000 60,000 NADPH + Oligos + Ligase (20min Rxn) 120,000 •Luminescent, Free of fluorescence interference 5.00 •HTS Formatted (96-, 384-, and 1536-well Plates) 4.00 R² = 0.998 NAD + Oligos + Ligase (20min Rxn) 100,000 80,000 3.00 60,000 40,000 •Universal: AMP and ADP Detection in the Presence or Absence of ATP as Substrate •Homogenous, Nonradioactive and Antibody free ADP (mM) NADH + Oligos + Ligase (20min Rxn) 160,000 100,000 ATP (mM) 6.00 NAD + Oligos + Ligase (20min Rxn) 180,000 mM 140,000 RLU 8.00 Nicodinamide specificity as Substrate in E. coli DNA Ligase (10µM Oligos Using AMP-Glo™) 12. Features of Promega Luminescent GloTM Technologies •Robust Assays (Z’>0.8) 2.00 40,000 20,000 20,000 0 1.00 0 0 0.5 1 1.5 2 E. coli DNA Ligase, U/Rxn/5µl 2.5 3 0 2 4 6 8 10 12 •Ultrasensitive (pmoles of nucleotides) Nicodinamides (µM) 0.00 HEK293 (10K cells/well) HEK293 (25K cells/well) CHO (10K cells/well) CHO (25K cells/well) •Stable Signal Kinase-Glo® measures the remaining ATP after a kinase Reaction www.promega.com For technical information: Said.Goueli@Promega.com