Jeremy E. Wilusz, Ph.D. Assistant Professor of Biochemistry and
advertisement

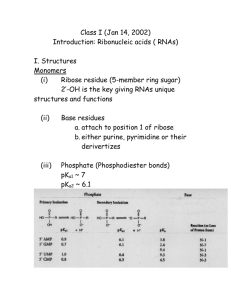
Jeremy E. Wilusz, Ph.D. Assistant Professor of Biochemistry and Biophysics (February 2014) B.S. Johns Hopkins University (2005) Ph.D. Watson School of Biological Sciences, Cold Spring Harbor Laboratory (2009) For Pubmed search: Wilusz je [author] DESCRIPTION OF RESEARCH INTERESTS: The sequencing of the human genome provided quite a surprise to many when it was determined that there are only ~20,000 protein-coding genes, representing less than 2% of the total genomic sequence. Since other less complex eukaryotes like the nematode C. elegans have a very similar number of genes, it quickly became clear that the developmental and physiological complexity of humans probably can not be solely explained by proteins. We now know that most of the human genome is transcribed, yielding a complex repertoire of RNAs that includes tens of thousands of individual noncoding RNAs with little or no proteincoding capacity. Among these are well-studied small RNAs, such as microRNAs, as well as many other classes of small and long transcripts whose functions and mechanisms of biogenesis are less clear – but likely no less important. This is because many of these poorly characterized RNAs exhibit cell type-specific expression or are associated with human diseases, including cancer and neurological disorders. Our goal is to characterize the mechanisms by which noncoding RNAs are generated, regulated, and function, thereby revealing novel fundamental insights into RNA biology and developing new methods to treat diseases. Much of our work has focused on the MALAT1 locus, which is over-expressed in many human cancers and produces a long nuclear-retained noncoding RNA as well as a tRNA-like cytoplasmic small RNA (known as mascRNA). Despite being an RNA polymerase II transcript, the 3’ end of MALAT1 is produced not by canonical cleavage/polyadenylation but instead by recognition and cleavage of the tRNA-like structure by RNase P. Mature MALAT1 thus lacks a poly(A) tail yet is expressed at a level higher than many proteincoding genes in vivo. We recently showed that the 3’ end of MALAT1 is protected from 3’-5’ exonucleases by a highly conserved triple helical structure. Surprisingly, when this structure is placed downstream from an open reading frame, the transcript is efficiently translated in vivo despite the lack of a poly(A) tail. This result challenges the common paradigm that long poly(A) tails are required for efficient protein synthesis and suggests that non-polyadenylated RNAs may produce functional peptides in vivo via mechanisms that are likely independent of poly(A) binding protein. To address these issues, we are currently elucidating the molecular mechanism by which a triple helix functions as a translational enhancer. In addition, we are developing approaches to identify additional triple helices that form across the transcriptome, thereby revealing new paradigms for how RNA structures regulate gene expression. Besides MALAT1, only a handful of long RNA polymerase II transcripts (e.g. histone mRNAs) have been clearly shown to be processed at their 3’ ends via non-canonical mechanisms. This is likely because nearly all previous studies characterizing the transcriptome have focused only on polyadenylated RNAs, thereby missing RNAs that lack a poly(A) tail. Nevertheless, it is becoming increasingly clear that nonpolyadenylated RNAs are much more common and play significantly greater regulatory roles in vivo than previously appreciated. For example, an abundant class of circular RNAs has recently been identified in human and mouse cells. Two of these circular RNAs clearly function as microRNA sponges, but it is largely unknown why all the other circular RNAs are produced or how the splicing machinery selects certain regions of the genome (and not others) to circularize. We aim to identify and characterize additional RNAs whose 3’ ends are generated via unexpected mechanisms, thereby revealing novel paradigms for how RNAs are processed and, most importantly, new classes of noncoding RNAs with important biological functions.