intro_classI - University of Windsor
advertisement
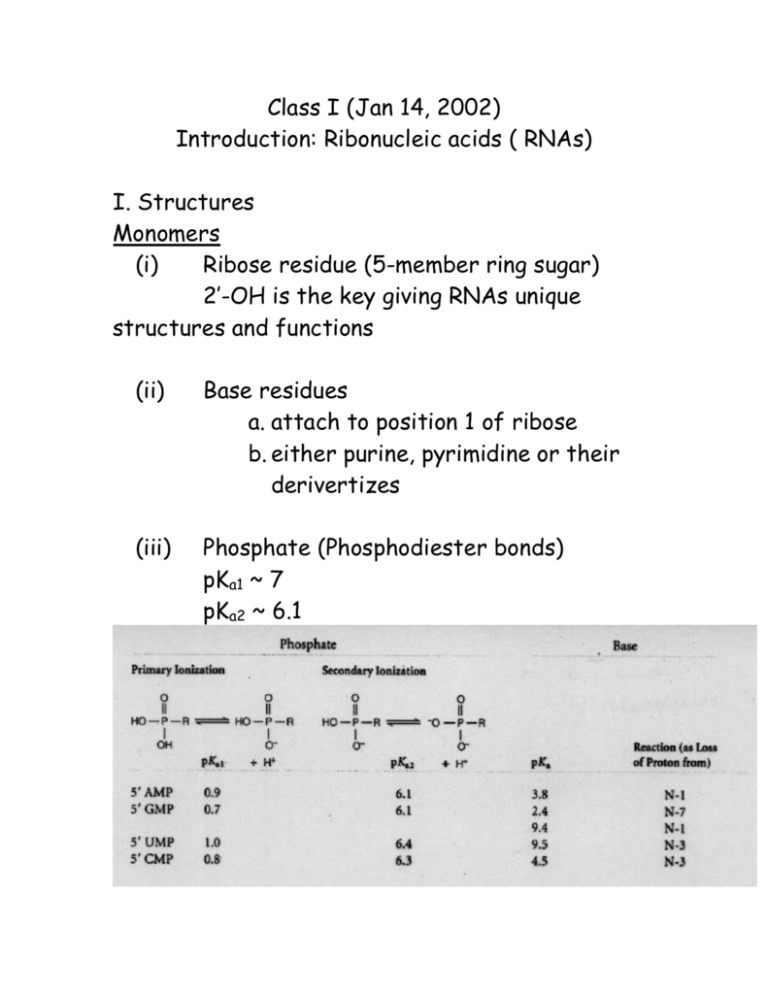
Class I (Jan 14, 2002) Introduction: Ribonucleic acids ( RNAs) I. Structures Monomers (i) Ribose residue (5-member ring sugar) 2’-OH is the key giving RNAs unique structures and functions (ii) Base residues a. attach to position 1 of ribose b. either purine, pyrimidine or their derivertizes (iii) Phosphate (Phosphodiester bonds) pKa1 ~ 7 pKa2 ~ 6.1 Polymers (i) Synthesis - RNA polymerases DNA Polymerases need primer 5’ 3’ extension 3’ 5’ nuclease RNA Polymerases no primer 5’ 3’ ext no nuclease - Chemical synthesis (ii) Nomenclature - 5’- and 3’ termini - Helicies (A form of double-stranded RNAs) - Base pairing, and stacking -Triple basepairs - Stem and loop structures Parameters of polynucleotide helices A Form Direction of helix rotation Right B Form Right Z Form Left Number of residues per turn (n) 11 10 12 (6 dimers) Rotation per residue (= 360o/n) 33o 36o -60o per dimer ~-30o per residue Rise in helix per residue (h) 0.255 nm 0.34 nm 0.37 nm Pitch of helix (=nh) 2.8 nm 3.4 nm 4.5 nm Accessibility poor good poor II. Functions of RNAs 2.1 Contemporary functions RNAs involve in.. a. Transcription e.g. RNAs are primers for DNA pol b. Translation e.g. mRNAs (templates), rRNAs and tRNAs (assemble of translation machinery). c. Regulation e.g. Ribosomal binding sequence, poly A tails, splice sites, etc. Contemporary functions: in concert with protein partners Ribonucleoprotein “RNP” 2.2 Prebiotic functions “When proteins are primitive and RNAs perform various functions from enzymes to genetic materials.” Prebiotic concept “RNA WORLD” (1986) Trigger: Ribozyme discovery Basic assumptions: (1) RNA replication assured genetic continuity (2) Basepair interaction had been a key in replication (3) Genetically encoded proteins were not involved as catalysts Evidence: 1. RNAs can perform catalytic activity. The discovery of two selfcatalytic RNAs, including RNase P and group I intron. RNAs can catalyze cleaving and ligation. 2. 3. 4. DNA synthesis requires ribonucleotide reductase to convert ribonucleotides to deoxyribonucleotides. Ribonucleotide reductase would probably be the primitive protein enzyme. RNA viruses use RNAs as genetic materials, templates to proteins. Ancient RNA viruses would have RNA telomere and RNA-telomerase. RNA molecules with peptide backbone or six member-ring sugar residues (pyranose) are nucleophilic, stable as templates for base-pairing, protein synthesis and etc. Genomic Tag Hypothesis (Weiner & Maizels, 1987) tRNA-like molecules can perform various functions : a. Replication of ss RNA viruses b. Replication of duplex DNA plasmids of fungal mitochondria c. Replication of retro virus d. Replication of chromosomal telomeres Hypothesis is that the tRNA-like structure evolved as 3’-terminal structures that tagged ancient RNA genomes for replication. Genomic Taq is the “Top half” of tRNA structure = minihelix The minihelix would be used as an initiation site for replication. In other word it is a simple telomere. Supportive documents found in present biological systems 1. RNase P is a ribonucleoprotein enzyme removing the 5’-leader from the tRNA precursors. Why tRNA should be processed? Possibly RNaseP would have been used to release functional RNAs from RNA genome. E. coli ‘s RNase P can function using only RNA component and process the 3’-terminal tRNA-like pseudoknot of turnip yellow mosaic virus. 2. Nucleotidyltransferase catalyzes the additing of CCA onto the 3’end of processed tRNA to ensure the presence of CCA. This enzyme is highly conserved. 3. Aminoacylation of tRNA (tRNA charging) is chemically similar to RNA polymerization. 4. Neurospora mitochondrial retroplasmids require RNA as intermediates. Similar phenomena are found in.. Is that possible that telomerase could have an internal genomic tag (CmAn motif)? Is telomerase a ribonucleoprotein enzyme? It is true at least in the case of Tetrahymena. Comparison between telomerase and retroplasmid Reverase transcriptase. Telomeraes: universal CmAn motif in eukaryotic nuclear chromosome ends 3’-CCA RNA motifs 5’ terminal motif (CmAn, RNA) (TnGm, DNA) Possible functions of tRNAs : Templates and Primers III. Research tools and techniques a. RNA isolation b. In vitro translation reaction c. Radioactive labelling d. Reverse transcription e. Gel electrophoresis f. Spectrofluorometric methods (Fluorescense energy transfer) g. Protection assays