pGFPuv - CMB Education
advertisement
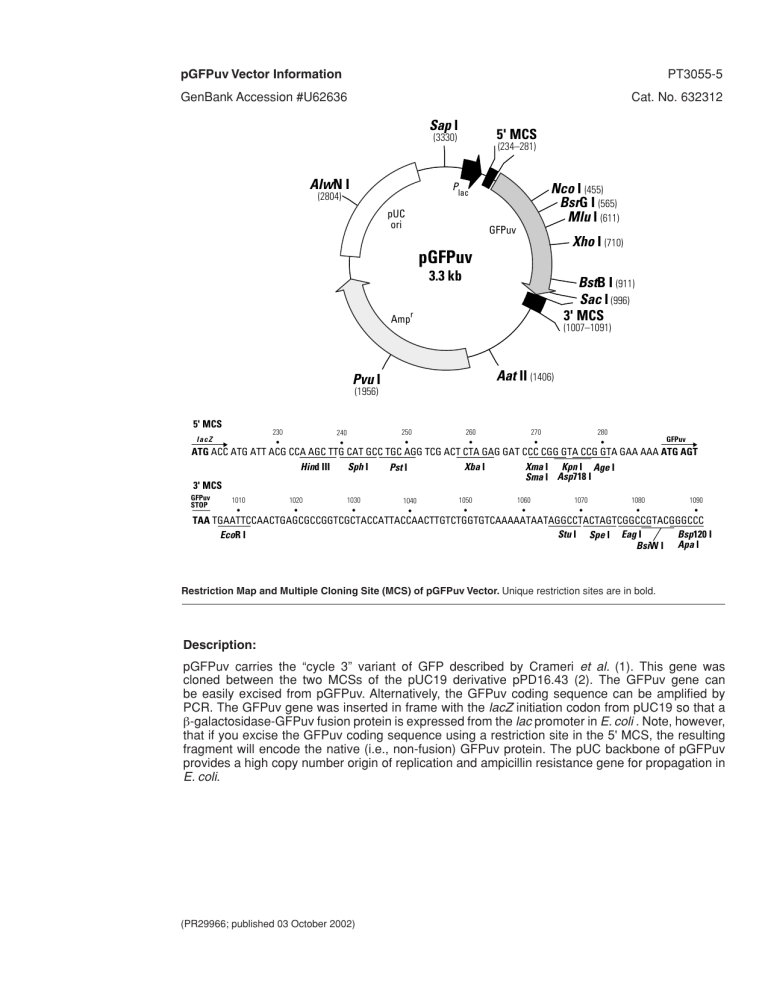
pGFPuv Vector Information PT3055-5 GenBank Accession #U62636 Cat. No. 632312 Sap I 5' MCS (3330) AlwN I P (2804) (234–281) Nco I (455) BsrG I (565) Mlu I (611) lac pUC ori GFPuv Xho I (710) pGFPuv 3.3 kb BstB I (911) Sac I (996) 3' MCS Ampr (1007–1091) Aat II (1406) Pvu I (1956) 5' MCS 230 • lacZ 250 • 240 • 260 • 270 • 280 • GFPuv ATG ACC ATG ATT ACG CCA AGC TTG CAT GCC TGC AGG TCG ACT CTA GAG GAT CCC CGG GTA CCG GTA GAA AAA ATG AGT Hind III Xba I Sph I Xma I Kpn I Age I Pst I Sma I Asp718 I 3' MCS GFPuv STOP 1010 • 1020 • 1030 • 1040 • 1050 • 1060 • 1070 • 1080 • 1090 • TAA TGAATTCCAACTGAGCGCCGGTCGCTACCATTACCAACTTGTCTGGTGTCAAAAATAATAGGCCTACTAGTCGGCCGTACGGGCCC Bsp10 I Stu I Spe I Eag I EcoR I BsiW I Apa I Restriction Map and Multiple Cloning Site (MCS) of pGFPuv Vector. Unique restriction sites are in bold. Description: pGFPuv carries the “cycle 3” variant of GFP described by Crameri et al. (1). This gene was cloned between the two MCSs of the pUC19 derivative pPD16.43 (2). The GFPuv gene can be easily excised from pGFPuv. Alternatively, the GFPuv coding sequence can be amplified by PCR. The GFPuv gene was inserted in frame with the lacZ initiation codon from pUC19 so that a β-galactosidase-GFPuv fusion protein is expressed from the lac promoter in E. coli . Note, however, that if you excise the GFPuv coding sequence using a restriction site in the 5' MCS, the resulting fragment will encode the native (i.e., non-fusion) GFPuv protein. The pUC backbone of pGFPuv provides a high copy number origin of replication and ampicillin resistance gene for propagation in E. coli. (PR29966; published 03 October 2002) pGFPuv Vector Information Location of features: • lac promoter: 95–178 CAP binding site: 111–124 –35 region: 143–148; –10 region: 167–172 Transcription start point: 179 lac operator: 179–199 • lacZ-GFPuv fusion protein expressed in E. coli Ribosome binding site: 206–209 Start codon (ATG): 217–219; Stop codon: 1003–1005 • 5' MCS: 234–281 • GFPuv gene Start codon (ATG): 289–291; Stop codon: 1003–1005 GFP chromophore: 481–489 wt GFP cDNA sequences (3): 289–454 Synthetic GFP gene with "cycle 3" mutations from pBAD-GFPuv (1): 455–1007 Cycle 3 mutation F99S (T→C): 584 Cycle 3 mutation M153T (T→C): 7Cycle 3 Cycle 3 mutation V163A (T→C): 776 Cycle 3 silent mutation in L137 (T→C): 699 Cycle 3 silent mutation in T225 (A→T): 963 Q80R mutation (A→G) (4): 527 Arg codons optimized for E. coli: R73 (AgA→CgT): 505–507, R96 (AgA→CgC): 574–576, R12(AgA→CgT): 652–654, R168 (AgA→CgC): 790–792, R1215 (AgA→CgT): 931–933 Silent mutations (CccA→TccG) creating BspE I site: 510 & 513 Silent mutation (A→G) creating Mlu I site: 612 Silent mutations (TtGgaA→CtCgaG) creating Xho I site: 709, 711 & 714 Silent mutations (AG→TC) creating BamH I site: 811–812 Silent mutation (C→G) creating Sal I site: 894 Silent mutations (ActA→GctC) creating Sac I site: 993 & 996 Silent mutation in S72 (A→C): 504 • 3' MCS: 107–1091 • Ampicilin resistance gene Promoter: –35 region: 1467–1472; –10 region: 1490–1495 Transcription start point: 1502 Ribosome binding site: 1525–1529 β-lactamase coding sequences: Start codon (ATG): 1537–1539; Stop codon: 2395–2397 β-lactamase signal peptide: 1537–1605 β-lactamase mature protein: 1606–2394 • pUC plasmid replication origin: 2545–3188 Primer Location: • GFP-N Sequencing Primer: 331–352 (Note: The GFP-C Sequencing Primer cannot be used with pGFPuv.) Propagation in E. coli: • Recommended host strain: JM109 or DH5α • Selectable marker: plasmid confers resistance to ampicillin (100 µg/ml) on E. coli hosts. • E. coli replication origin: pUC • Copy number: ≈500 • Plasmid incompatibility group: pMB1/ColE1 References: 1. 2. 3. 4. Crameri, A., et al. (1996) Nature Biotechnol. 14:315–319. Fire, A., et al. (1990) Gene 93:189–198. Prasher, D. C., et al. (1992) Gene 111:229–233. Chalfie, M., et al. (1994) Science 263:802–805. Clontech Laboratories, Inc. www.clontech.com Protocol No. PT3055-5 Version No. PR29966 Fri, Jul 12, 1996 Digest 1 of pGFPuv: 3337 bases (circular) Page 1 AseI PvuII | | AGCGCCCAATACGCAAACCGCCTCTCCCCGCGCGTTGGCCGATTCATTAATGCAGCTGGCACGACAGGTT TCGCGGGTTATGCGTTTGGCGGAGAGGGGCGCGCAACCGGCTAAGTAATTACGTCGACCGTGCTGTCCAA 10 20 30 40 50 60 70 AseI | TCCCGACTGGAAAGCGGGCAGTGAGCGCAACGCAATTAATGTGAGTTAGCTCACTCATTAGGCACCCCAG AGGGCTGACCTTTCGCCCGTCACTCGCGTTGCGTTAATTACACTCAATCGAGTGAGTAATCCGTGGGGTC 80 90 100 110 120 130 140 BsrBI | GCTTTACACTTTATGCTTCCGGCTCGTATGTTGTGTGGAATTGTGAGCGGATAACAATTTCACACAGGAA CGAAATGTGAAATACGAAGGCCGAGCATACAACACACCTTAACACTCGCCTATTGTTAAAGTGTGTCCTT 150 160 170 180 190 200 210 Sal I Asp718 I BspMI PstI XbaI XmaI KpnI HindIII SphI Sse8387 I BamHI SmaI AgeI | | | | | | | | | | || ACAGCTATGACCATGATTACGCCAAGCTTGCATGCCTGCAGGTCGACTCTAGAGGATCCCCGGGTACCGG TGTCGATACTGGTACTAATGCGGTTCGAACGTACGGACGTCCAGCTGAGATCTCCTAGGGGCCCATGGCC 220 230 240 250 260 270 280 BpmI | TAGAAAAAATGAGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCCAATTCTTGTTGAATTAGATGGTGA ATCTTTTTTACTCATTTCCTCTTCTTGAAAAGTGACCTCAACAGGGTTAAGAACAACTTAATCTACCACT 290 300 310 320 330 340 350 TGTTAATGGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATACGGAAAACTTACCCTT ACAATTACCCGTGTTTAAAAGACAGTCACCTCTCCCACTTCCACTACGTTGTATGCCTTTTGAATGGGAA 360 370 380 390 400 410 420 NcoI MscI | | AAATTTATTTGCACTACTGGAAAACTACCTGTTCCATGGCCAACACTTGTCACTACTTTCTCTTATGGTG TTTAAATAAACGTGATGACCTTTTGATGGACAAGGTACCGGTTGTGAACAGTGATGAAAGAGAATACCAC 430 440 450 460 470 480 490 BspEI NdeI | | TTCAATGCTTTTCCCGTTATCCGGATCATATGAAACGGCATGACTTTTTCAAGAGTGCCATGCCCGAAGG AAGTTACGAAAAGGGCAATAGGCCTAGTATACTTTGCCGTACTGAAAAAGTTCTCACGGTACGGGCTTCC 500 510 520 530 540 550 560 BsrGI MluI | | TTATGTACAGGAACGCACTATATCTTTCAAAGATGACGGGAACTACAAGACGCGTGCTGAAGTCAAGTTT AATACATGTCCTTGCGTGATATAGAAAGTTTCTACTGCCCTTGATGTTCTGCGCACGACTTCAGTTCAAA 570 580 590 600 610 620 630







