Microcephaly info sheet-12.14.15
advertisement
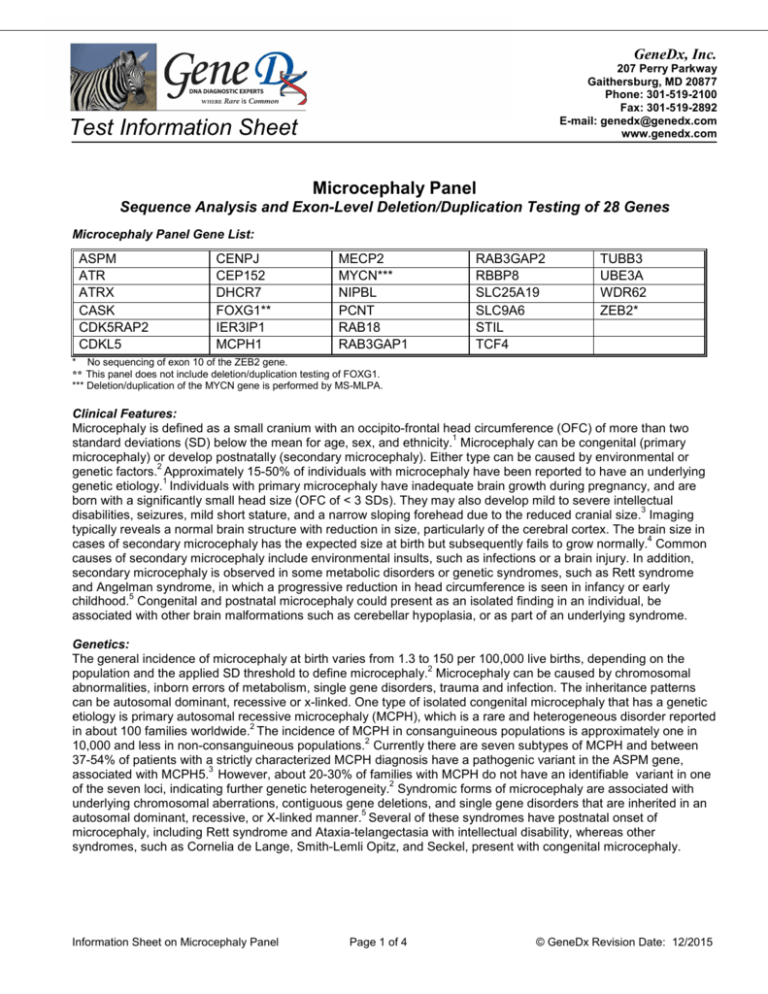
GeneDx, Inc. 207 Perry Parkway Gaithersburg, MD 20877 Phone: 301-519-2100 Fax: 301-519-2892 E-mail: genedx@genedx.com www.genedx.com Test Information Sheet Microcephaly Panel Sequence Analysis and Exon-Level Deletion/Duplication Testing of 28 Genes Microcephaly Panel Gene List: ASPM ATR ATRX CASK CDK5RAP2 CDKL5 CENPJ CEP152 DHCR7 FOXG1** IER3IP1 MCPH1 MECP2 MYCN*** NIPBL PCNT RAB18 RAB3GAP1 RAB3GAP2 RBBP8 SLC25A19 SLC9A6 STIL TCF4 TUBB3 UBE3A WDR62 ZEB2* * No sequencing of exon 10 of the ZEB2 gene. ** This panel does not include deletion/duplication testing of FOXG1. *** Deletion/duplication of the MYCN gene is performed by MS-MLPA. Clinical Features: Microcephaly is defined as a small cranium with an occipito-frontal head circumference (OFC) of more than two 1 standard deviations (SD) below the mean for age, sex, and ethnicity. Microcephaly can be congenital (primary microcephaly) or develop postnatally (secondary microcephaly). Either type can be caused by environmental or 2 genetic factors. Approximately 15-50% of individuals with microcephaly have been reported to have an underlying 1 genetic etiology. Individuals with primary microcephaly have inadequate brain growth during pregnancy, and are born with a significantly small head size (OFC of < 3 SDs). They may also develop mild to severe intellectual 3 disabilities, seizures, mild short stature, and a narrow sloping forehead due to the reduced cranial size. Imaging typically reveals a normal brain structure with reduction in size, particularly of the cerebral cortex. The brain size in 4 cases of secondary microcephaly has the expected size at birth but subsequently fails to grow normally. Common causes of secondary microcephaly include environmental insults, such as infections or a brain injury. In addition, secondary microcephaly is observed in some metabolic disorders or genetic syndromes, such as Rett syndrome and Angelman syndrome, in which a progressive reduction in head circumference is seen in infancy or early 5 childhood. Congenital and postnatal microcephaly could present as an isolated finding in an individual, be associated with other brain malformations such as cerebellar hypoplasia, or as part of an underlying syndrome. Genetics: The general incidence of microcephaly at birth varies from 1.3 to 150 per 100,000 live births, depending on the 2 population and the applied SD threshold to define microcephaly. Microcephaly can be caused by chromosomal abnormalities, inborn errors of metabolism, single gene disorders, trauma and infection. The inheritance patterns can be autosomal dominant, recessive or x-linked. One type of isolated congenital microcephaly that has a genetic etiology is primary autosomal recessive microcephaly (MCPH), which is a rare and heterogeneous disorder reported 2 in about 100 families worldwide. The incidence of MCPH in consanguineous populations is approximately one in 2 10,000 and less in non-consanguineous populations. Currently there are seven subtypes of MCPH and between 37-54% of patients with a strictly characterized MCPH diagnosis have a pathogenic variant in the ASPM gene, 3 associated with MCPH5. However, about 20-30% of families with MCPH do not have an identifiable variant in one 2 of the seven loci, indicating further genetic heterogeneity. Syndromic forms of microcephaly are associated with underlying chromosomal aberrations, contiguous gene deletions, and single gene disorders that are inherited in an 5 autosomal dominant, recessive, or X-linked manner. Several of these syndromes have postnatal onset of microcephaly, including Rett syndrome and Ataxia-telangectasia with intellectual disability, whereas other syndromes, such as Cornelia de Lange, Smith-Lemli Opitz, and Seckel, present with congenital microcephaly. Information Sheet on Microcephaly Panel Page 1 of 4 © GeneDx Revision Date: 12/2015 Reason for Referral: 1. Molecular confirmation of a clinical diagnosis. 2. Establish the type of microcephaly and cause in order to provide information regarding prognosis, management and recurrence risk. 3. Testing of at-risk relatives for specific known variant(s) previously identified in an affected family member. 4. Prenatal diagnosis for known familial pathogenic variant(s) in at-risk pregnancies. Methods: Using genomic DNA, coding exons and flanking splice junctions of the genes on this panel are enriched using a proprietary targeted capture method developed by GeneDx. The products are sequenced on an Illumina instrument using paired end reads. The sequence data is aligned to reference sequences based on human genome build GRCh37/UCSC hg19. Sanger sequencing is used to compensate for low coverage and refractory amplifications. Concurrently, multiplex ligation-dependent probe amplification (MLPA) of the MYCN gene and targeted array CGH analysis with exon-level resolution is performed to evaluate for a deletion or duplication of one or more exons for most of the genes included on the panel. If indicated, MLPA of the FOXG1 gene is available as a separate test (test code 904). The presence of any potentially disease-associated sequence variant(s) or copy number alteration(s) is confirmed by dideoxy DNA sequence analysis or quantitative PCR, respectively, or by other appropriate methods. Test Sensitivity: The clinical sensitivity of sequencing and deletion/duplication analysis of the genes included in this panel depends on the patient’s clinical phenotype. Specific information about the diagnostic yield for each gene in selected populations is summarized in the following table. The technical sensitivity of the sequencing test is estimated to be 98%. Deletions involving more than 20 bp and insertions involving more than 10 bp are not reliably detected by the sequencing methodology, and deletions or duplications of less than 500 bp are not reliably detected by array CGH. Note that small sections of a few genes have inherent sequence properties that result in suboptimal data and variants in those regions may not be reliably identified. Specimen Requirements and Shipping/Handling: • Blood: Whole blood in EDTA. Adults: 8-10 ml; Children: 4 ml; Infants: 2 ml. Ship blood overnight at ambient temperature, using a cool pack in hot weather. Blood specimens may be refrigerated for up to 7 days prior to shipping. • Oral Rinse: Use GeneDx kit only; follow instructions at http://www.genedx.com/order-a-test/specimenrequirements/. • Extracted DNA: Outside DNA is discouraged; however, high quality extracted DNA can be accepted. This test requires a minimum of 20 ug of DNA at a concentration of 50 ng/ul of DNA with a minimum volume of 400 ul. • Other specimens: Buccal brushes and paraffin embedded tissue CANNOT be used for this testing. • Prenatal Diagnosis (for specific known mutation(s) or deletion(s) only): For prenatal testing for a known pathogenic variant in a gene on this panel, please refer to the specimen requirements table on our website at: http://ww.genedx.com/test-catalog/prenatal/. Ship specimen overnight at ambient temperature, using a cool pack in hot weather. Test 689 9011/9012/9013 441 902 Description Microcephaly Panel DNA sequencing of a relative for one/two/three specific known variant(s) Deletion/duplication testing of a relative for specific known deletion or duplication Prenatal diagnosis for specific known pathogenic variant(s) References: 1. Ashwal et al., (2009) Neurology 73: 887-897. 2. Kaindl et al., (2010) Progress in Neurobiology 90:363-383. 3. Passemard et al., (2009) Primary Autosomal Recessive Microcephaly. In : GeneReviews at GeneTests: Medical Genetics Information Resource (database online). Copyright, University of Washington, Seattle. 1997-2010. Available at http://www.genetests.org. 4. Woods CG (2004) Current Opinion in Neurobiology 14:112-117. 5. Abuelo, D (2007) Semin Pediatr Neurol 14:118-127. 6. Thornton GK and Woods CG (2009) Trends in Genetics 25(11):501-510. Information Sheet on Microcephaly Panel Page 2 of 4 © GeneDx Revision Date: 12/2015 Genes Associated with Non-syndromic Microcephaly Disorder Gene MCPH1 WDR62 (MCPH2) CDK5RAP2 (MCPH3) Primary Microcephaly (Types 1-7) CEP152 (MCPH9 ; previously MCPH4) ASPM (MCPH5) CENPJ (MCPH6) STIL (MCPH7) Protein Microcephalin Inh. AR Diagnostic Yield in Selected Population(s) <5% of MCPH1 WD repeat-containing protein 62 AR Rare2 Cyclin-dependent kinase 5 regulatory subunit associated protein 2 AR <5% of MCPH1 Centrosomal protein of 152 kDa AR Rare for MCPH3 Rare in Seckel syndrome4 AR 37-54% of MCPH1 AR <5% of MCPH1 Rare in Seckel syndrome5 AR <5% of MCPH1 Abnormal spindle-like, microcephaly-associated protein Centromeric associated protein J SCL/TAL1-interrupting locus protein Alpha-Thalassemia Intellectual Disability Syndrome ATRX Transcriptional regulator ATRX XL ~25% in ATRX6 Amish Lethal Microcephaly SLC25A19 Nuclear mitochondrial deoxynucleotide carrier AR 100% in Old Order Amish population7 Otherwise rare Complex Cortical Dysplasia, with Other Brain Malformations (CDCBM) TUBB3 Tubulin beta-3 AD Rare8 Mental Retardation and Microcephaly with Pontine and Cerebellar Hypoplasia CASK Calcium/calmodulin-dependent serine protein kinase XL ~4% in cerebellar hypoplasia and intellectual 9-11 disability References: 1. Passemard et al., (2009) Primary Autosomal Recessive Microcephaly. In: GeneReviews at GeneTests: Medical Genetics Information Resource (database online). Copyright, University of Washington, Seattle. 1997-2010. Available at http://www.genetests.org. 2. Yu et al., (2010) Nat Genet 42(11):1015-20. 3. Guernsey et al., (2010) Am J Hum Genet 87(1):40-51. 4. Kalay et al., (2011) Nat Genet 43(1):23-26. 5. Al-Dosari et al., (2010) J Med Genet 47:411-414. 6. Badens et al., (2006) Clin Genet 70(1):57-62. 7. Dunckelmann et al., (2000) Neuropediatrics 31:35-8. 8. Poirier et al., (2010) Hum Mol Genet 19:4462–4473. 9. Najm et al., (2008) Nat Genet Sep;40(9):1065-7. 10. Hackett et al., (2010) Eur J Hum Genet May;18(5):544-52. 11. Burglen et al., (2012) Orphanet J Rare Dis Mar 27;7:18. Information Sheet on Microcephaly Panel Page 3 of 4 © GeneDx Revision Date: 12/2015 Genes Associated with Syndromic Microcephaly Syndrome Gene Protein Inh. Diagnostic Yield in Selected Populations 88% females with Rett syndrome1 2-8% females with atypical Rett syndrome2,3 MECP2 Methyl CpG binding protein 2 XL CDKL5 Cyclin-dependent kinase-like 5 XL FOXG1** Forkhead box protein G1 AD ~1% Rett syndrome overall4 25% congenital variant of Rett5 UBE3A Ubiquitin protein ligase E3A AD 11% Angelman syndrome6 SLC9A6 Sodium/hydrogen exchanger 6 XL 6% Angelman-like syndrome7 TCF4 Transcription factor 4 AD 36% Pitt-Hopkins syndrome (PHS)8 2% Angelman syndrome8 Cornelia de Lange syndrome NIPBL Delangin AD ~37-47% Cornelia de lange syndrome (CdLS)9-11 Feingold syndrome MYCN*** N-myc proto-oncogene protein AD ~65-75% Feingold syndrome12-14 Mowat-Wilson syndrome ZEB2* Zinc finger E-box-binding homeobox 2 AD 95% Mowat Wilson syndrome15-16 Microcephaly with simplified gyral pattern, epilepsy and permanent neonatal diabetes syndrome (MEDS) IER3IP1 Immediate-early response 3interacting protein 1 AR Unknown17-18 ATR Ataxia-telangiectasia and Rad3related protein AR Unknown19 PCNT Pericentrin AR ~30% in Seckel syndrome20 100% in Microcephalic Osteodysplastic Primoridal Dwarfism type II (MOPDII)20,21 RBBP8 CTBP interacting protein AR Unknown22,23 Smith-Lemli Opitz syndrome DHCR7 7-dehydrocholesterol reductase AR ~95% Smith Lemli Opitz (one or both variants)24 Warburg Micro Syndrome RAB18 RAS-associated protein AR Rare25 AR ~67% Warburg micro syndrome26 AR Unknown in Warburg micro syndrome27 Unknown in Martsolf syndrome28 Typical/Atypical Rett syndromes Angelman/ Angelman-like/ Pitt-Hopkins syndromes Seckel syndrome RAB3GAP1 RAB3GAP2 Rab3 GTPase-activating protein (catalytic subunit) Rab3 GTPase-activating protein (non-catalytic subunit) * No sequencing of exon 10 of the ZEB2 gene. ** This panel does not include deletion/duplication testing of FOXG1. ***Deletion/duplication of the MYCN gene is performed by MS-MLPA. References: 1. Li et al., (2006) J Hum Genet 52:38-47. 2. Tao et al., (2004) Am J Hum Genet 75:1149-1154. 3. Rosas-Vargas et al., (2008) J Med Genet 45:172-178. 4. Bahi-Buisson et al., (2010) 11:241-249. 5. Mencarelli et al., (2010) J Med Genet 47:49-53. 6. Lossie et al., (2001) J Med Genet 38(12):834-845. 7. Gillfillan et al., (2008) Am J Hum Genet 82:1003-1010. 8. de Pontual et al., (2009) Hum Mutat 30:669-676. 9. Pie et al., (2010) Am J Med Genet 152A(4):924-929. 10. Selicorni et al., (2007) Clin Genet 72:98-108. 11. Gillis et al. (2004) Am J Hum Genet 75:610-623. 12. Marcelis et al., (2008) Human Mutation 29(9):1125-1132. 13. Van Bokhoven et al., (2005) Nat Genet 37:465-467. 14. Teszas et al., (2006) Am J Med Genet 140A:2254-56. 15. Wilson et al., (2003) Am J Med Genet 119A:257-265. 16. Zweier et al., (2005) Eur J Med Genet 48:97-111. 17. DeWit et al., (2006) Neurogenetics 7:259-263. 18. Poulton et al. (2011) Am J Hum Genet 89: 265-276. 19. O’Driscoll et al., (2003) Nature Genetics 33: 495-501. 20. Willems et al., (2010) J Med Genet 47: 797-802. 21. Rauch et al., (2008) Science 319:816-819. 22. Borglum et al., (2001) Eur J Hum Genet 9:753-757. 23. Qvist et al., (2011) PLoS Genet 7:e1002310. 24. Witsch-Baumgartner et al. (2000) Am J Hum Genet 66:402-412. 25. Bern et al., (2011) Am J Hum Genet 129:45-50. 26. Aligianis et al., (2005) Nat Genet 37:211-223. 27. Borck et al., (2011) Hum Genet 129:45-50. 28. Aligianis et al., (2006) Am J Hum Genet 78:702-7. Information Sheet on Microcephaly Panel Page 4 of 4 © GeneDx Revision Date: 12/2015