BBAMCR-16874; No. of pages: 16; 4C: 2, 5, 7
Biochimica et Biophysica Acta xxx (2013) xxx–xxx
Contents lists available at SciVerse ScienceDirect
Biochimica et Biophysica Acta
journal homepage: www.elsevier.com/locate/bbamcr
Review
Organization and function of membrane contact sites☆
Sebastian C.J. Helle 1, Gil Kanfer 1, Katja Kolar 1, Alexander Lang 1, Agnès H. Michel 1, Benoît Kornmann ⁎
Institute of Biochemistry, ETH Zürich, HPM G16 Schafmattstrasse, 18 8093 Zürich Switzerland
a r t i c l e
i n f o
Article history:
Received 22 November 2012
Received in revised form 18 January 2013
Accepted 21 January 2013
Available online xxxx
Keywords:
Membrane contact site
Organelle
Endoplasmic reticulum
Calcium
Lipid
Interorganelle communication
a b s t r a c t
Membrane-bound organelles are a wonderful evolutionary acquisition of the eukaryotic cell, allowing the
segregation of sometimes incompatible biochemical reactions into specific compartments with tailored microenvironments. On the flip side, these isolating membranes that crowd the interior of the cell, constitute
a hindrance to the diffusion of metabolites and information to all corners of the cell. To ensure coordination
of cellular activities, cells use a network of contact sites between the membranes of different organelles.
These membrane contact sites (MCSs) are domains where two membranes come to close proximity, typically
less than 30 nm. Such contacts create microdomains that favor exchange between two organelles. MCSs are
established and maintained in durable or transient states by tethering structures, which keep the two
membranes in proximity, but fusion between the membranes does not take place. Since the endoplasmic
reticulum (ER) is the most extensive cellular membrane network, it is thus not surprising to find the ER
involved in most MCSs within the cell. The ER contacts diverse compartments such as mitochondria,
lysosomes, lipid droplets, the Golgi apparatus, endosomes and the plasma membrane. In this review, we
will focus on the common organizing principles underlying the many MCSs found between the ER and virtually all compartments of the cell, and on how the ER establishes a network of MCSs for the trafficking of vital
metabolites and information. This article is part of a Special Issue entitled: Functional and structural diversity
of endoplasmic reticulum.
© 2013 Elsevier B.V. All rights reserved.
1. Functions of MCSs
MCSs display a wide variety of functions including, but not limited
to regulation of immune response, apoptosis, organelle dynamics and
trafficking. At the molecular level, the function of MCSs most often
involves calcium (Ca2+) and/or lipid exchange between two compartments. Trafficking of both Ca2+ and lipids must be governed by strict
rules. In the case of Ca 2+, tight coordination is required because it is
a very potent signaling molecule that acts locally, and which can be
toxic when deregulated (reviewed in [1]), and in the case of lipids
because their variety contributes to determining the identity and
physicochemical properties of all membrane-bound compartments
(reviewed in [2]). Ca2+ and lipids also have in common diffusionlimited properties. For Ca 2+, diffusion is limited by the abundance of
Ca2+-binding activities in all compartments of the cell, while diffusion
of lipids is limited by their insolubility in most of the aqueous cellular
volume. MCSs allow localized and targeted exchange of both Ca2+
and lipids to coordinate homeostasis across the many compartments
of the cell. Since the ER is both the main site of lipid synthesis and
the main cellular Ca2+ store, it is not surprising that MCSs involving
☆ This article is part of a Special Issue entitled: Functional and structural diversity of
endoplasmic reticulum.
⁎ Corresponding author. Tel.: +41 44 633 65 75.
E-mail address: benoit.kornmann@bc.biol.ethz.ch (B. Kornmann).
1
Equal participation.
the ER provide an important function in both of these exchange
reactions.
1.1. Calcium signaling
The ER [Ca 2+] is in the millimolar range, while that of the cytosol
is nanomolar (reviewed in [1]). This difference is established by
Sarcoplasmic/Endoplasmic Reticulum Ca 2+ ATPases (SERCA), which
pump Ca 2+ into the ER lumen, and by Plasma Membrane Ca 2+
ATPases (PMCA), which pump it out of the cell (Fig. 1B). This gradient
is used in signaling events whereby the opening of ER Ca 2+ channels
(the Ryanodine Receptors, RyR and the Inositol-1,4,5-triphosphate
Receptor, IP3R) causes a transient surge in cytosolic [Ca 2+], triggering
a number of downstream events by activating Ca 2+-binding proteins.
Ca 2 + signaling governs processes as diverse as memory, vision,
fertilization, muscle contraction, proliferation, cell migration, immune
response and transcription. Ca2+ does not only signal to the cytosol,
but also to other compartments. Intracellular Ca2+ diffusion is slowed
by the presence of many Ca2+-binding activities. This slow diffusion
maintains steep local [Ca2+] gradients and non-equilibrium conditions,
which are best illustrated by such spectacular phenomena as Ca2+
oscillations and Ca 2+ waves [1]. On the small scale, slow Ca2+ diffusion
promotes the formation of cytosolic Ca 2+ microdomains, where the
[Ca2+] differs from that of the bulk cytosol. Microdomains of this
kind are often created at MCSs, which coordinate vast movements of
the Ca2+ pool across different compartments.
0167-4889/$ – see front matter © 2013 Elsevier B.V. All rights reserved.
http://dx.doi.org/10.1016/j.bbamcr.2013.01.028
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
2
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
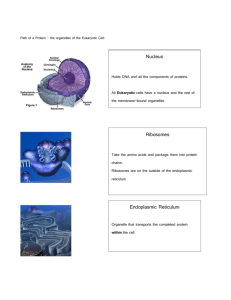
Fig 1. MCSs and Ca2+ homeostasis. A) Organization of the junctional membrane complexes in striated cardiac and skeletal muscles. DHPR (yellow) is found in T-tubules, which
invaginate from the cell surface into the cell interior. DHPR gates the opening of RyR (red) in cardiac cells by increasing the local [Ca2+] (Ca2+-gated Ca2+ release), in skeletal
cells by physically contacting them across the cytosolic space between the SR and the PM (allosteric coupling). B) Organization of Ca2+ microdomains at MCSs. The blue color represents the local [Ca2+]. A steep [Ca2+] gradient is established by constitutive Ca2+ pumping performed by SERCA (red) towards the ER lumen, and PMCA (yellow) towards cell
exterior. ER–PM MCSs allow direct ER Ca2+ refilling via STIM/Orai-dependent (purple/green) SOCE with minimal Ca2+ escape towards the bulk cytosol. ER–mitochondria MCSs
allow direct Ca2+ exchange from the ER channel IP3R (tan) to the mitochondrial uptake machinery VDAC (blue) and MCU (green).
1.1.1. Excitation–contraction in muscle cells
A classical contact site example is the spatial arrangement of the
muscle ER (called Sarcoplasmic Reticulum, SR) and the PlasmaMembrane (PM), and its role in generating the signal that results in
muscle contraction [3]. Muscle contraction is triggered by a massive
influx of Ca 2+ into the cytosol, which activates myosin movement
along actin filaments in the sarcomeres (Fig. 1A). This Ca 2+ influx
is the result of the synergistic activation of two mechanisms:
1) the opening of voltage-gated PM Ca 2+ channels, such as the
dihydropyridine receptor (DHPR), which respond to a change in PM
potential originating at the neuromuscular synapse, and 2) the synchronous opening of the main SR Ca 2+ channel RyR (reviewed in
[4]). The synchronization of both events is ensured by their physical
coupling at MCSs between the SR and the PM. The myocyte PM is
invaginated in between sarcomeres. These invaginations create transverse tubules (T-tubules), which project into the center of the cell.
These T-tubules are closely bracketed by SR tubules, forming characteristic junctional membrane complexes (JMC) [5], referred to as
dyads, where one T-tubule associates with one tubule of SR, or triads,
where one T-tubule is bracketed by two tubules of SR (Fig. 1A). RyRs
are found on the SR tubules while DHPRs are on the opposing
T-tubules. It is this proximity between RyR and DHPR that allows
coupled gating of both channels. Interestingly, although the spatial
proximity between both types of Ca 2+ channels is conserved, the
way DHPR “talks” to RyR depends on the muscle type. In cardiac muscle, myocardial RyR (RyR2) senses the Ca 2+ emanating from DHPR.
Ensuing RyR opening is therefore part of a positive feedback loop
[6]. In non-pulsatile skeletal muscles, however, DHPR is thought to directly open skeletal muscle RyR (RyR1) through a physical connection
between both channels [7,8]. According to this model, a voltagedependent change in DHPR conformation is transmitted to RyR1 via
the C-terminus of DHPR cytoplasmic β1a subunit. This subunit of
DHPR bridges the cytosolic gap in between T-tubule and SR to directly
bind RyR1 and gate its opening [9]. In both cases, close apposition of
SR and PM at MCSs allows the coordination of cellular events, which
permits rapid and precise muscular contraction.
1.1.2. The store-operated Ca 2+ entry
1.1.2.1. Function. Ca 2+ signaling is not restricted to excitable muscle
cells. Non-excitable cells also use Ca 2+ signaling in important signaling cascades, and they also use MCSs to coordinate these cascades
across the many compartments of the cell.
The ER lumen is the site of numerous fundamental calciumdependent processes, such as protein folding and quality control
[10]. After a discharge caused by opening of IP3R – the main Ca 2+
channel in the ER of non-excitable cells – it is essential that the luminal [Ca 2+] returns rapidly to its normal level. This is ensured by an
important cellular mechanism, which triggers the entry of Ca 2+
ions into the cytosol from the extracellular space in conditions
where the ER Ca 2+ store is depleted.
This Store-Operated Ca 2+ Entry (SOCE) is not only important for
ER function, but also for the proper execution of Ca 2+-dependent
signaling cascades. By constantly refilling the ER, SOCE allows to sustain elevated cytosolic [Ca 2+] during extended periods of times. For
instance, SOCE is required for many immune functions, such as
T-lymphocyte activation and proliferation (reviewed in [11]).
1.1.2.2. SOCE mechanism. How does a Ca2+ channel situated in the PM
know that the [Ca 2+] in the ER lumen is low? This question has kept
researchers busy for decades, until RNAi screens revealed an astoundingly simple device for membrane–membrane communication, made
of only two central components: 1) the sensor Stromal Interaction
Molecule (STIM) [12,13] and 2) the effector PM Ca 2+ channel Orai
[14–18]. STIM is a type I transmembrane ER protein. It senses luminal
[Ca2+] via its luminal N-terminus, which bears Ca2+-binding EF-hand
motifs [12]. Ca2+ depletion in the lumen causes the unfolding of the
EF-hands and allows the formation of STIM oligomers [19–21]. Oligomerization is accompanied by a conformational change in the Cterminal cytosolic moiety [22], exposing a polybasic tract of aminoacids
near the C-terminus. The polybasic tract can bind to acidic phosphoinositides found in the PM [19,22–24]. As a result, the localization
of STIM, which is rather homogenous in the ER in Ca2+ replete condition [12,19,25], becomes restricted to a few structures (“puncta”) upon
ER Ca 2+ depletion. Examination using TIRF microscopy (see Box 3)
indicates that these puncta are MCSs between the ER and the PM
[12,25–30]. At these ER–PM MCSs, STIM can recruit the Ca 2+-specific
channel Orai and gate it by direct physical interaction [31–35]. This
system is so simple that it can be recapitulated in vitro. Recombinant
STIM cytosolic fragments bind to and gate Orai channels produced in
yeast [36]. As yeast does not have any of the SOCE components, it is
very likely that this gating is direct.
1.1.2.3. SOCE and MCSs: how?. What recruits STIM to the MCSs? A likely model is that in resting conditions, STIM diffuses passively in the
plane of the ER membrane, and that upon stimulation, the exposure
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
of its phosphoinositide-binding polybasic domain causes STIM to be
trapped at preexisting ER–PM MCSs [25]. The examination of cycles
of SOCE activation/deactivation by TIRF microscopy reveals that
STIM puncta form repeatedly at the same location, suggesting that
the MCSs are indeed preexisting and are maintained between activation cycle [37]. Moreover, consistent with passive diffusion, formation
and dissociation of STIM oligomers, as well as its translocation to and
from puncta, do not require ATP [38]. The structures that establish
and maintain these preexisting MCSs are not known. However, the
extent of ER–PM connection is affected by SOCE activation or STIM
overexpression, suggesting that, on the contrary, STIM creates the
MCS that it will use to gate Orai [25,39,40]. It is possible that the
interaction of STIM with PM lipids and with Orai adds a tethering
force to preexisting MCSs, thereby enlarging them (see Section 2.3
and Box 1).
1.1.2.4. SOCE and MCSs: why?. Why is SOCE integrated at ER–PM
MCSs? By allowing the sensor in the ER to directly interact with the
effector in the PM, MCSs reduce the number of necessary players to
the strict minimum. But the extraordinary complexity of most cellular
signaling events shows that simplicity is hardly a selecting force. Instead, it is possible that integrating both SOCE sensor and effector at
MCSs fulfills other functions. Having PM Ca 2+ channels opened at
sites of contact with the ER may allow Ca 2+ to flow directly from
the extracellular space to the ER lumen, bypassing the bulk cytosol
(Fig. 1B), much like in the case of ER–mitochondrial Ca 2+ exchange
(see Section 1.1.3). This model finds supporting evidence: SERCA inhibitors are usually used to induce a full-blown SOCE that is not mitigated by ER refilling. In such conditions, SOCE activation causes a
massive increase in cytosolic [Ca 2+], but when SOCE is activated
without affecting SERCA, only a transient modest rise in cytosolic
[Ca 2+] is observed [41]. ER refilling can even take place without any
detectable cytosolic [Ca 2+] rise, provided that SOCE is slowed by a
mild knockdown of STIM or by limiting extracellular [Ca 2+]. This suggests that the tight coupling between PM opening and ER Ca 2+
pumping allows Ca 2+ to directly transfer from the extracellular
space to the ER lumen before it can diffuse into the bulk cytosol
(Fig. 1B) [42]. To achieve this tight coupling, the SERCA may directly
interact with STIM, a notion supported by co-immunoprecipitation
experiments [43].
Thus, integrating the ER [Ca 2+] signal at ER–PM MCSs may not
only be a way to shorten the signal transduction pathway, but it
may also be a way to minimize cytosolic [Ca 2+] perturbations while
refilling the ER pool.
3
Box 1
Static or dynamic?
Very dynamic processes happen at membrane contact sites,
begging the question of whether these are themselves static or
dynamic structures. In a growing yeast cell, the vast majority
of the cortical ER (i.e. the part of the ER that is not the nuclear envelope) is found in direct proximity with the plasma membrane
throughout cell life, suggesting that tethering is durable. ER–
mitochondria connections also appear to be stable structures.
ERMES complexes can be imaged for minutes in yeast [107]
and the dynamics of ER and mitochondria are coupled in the
lamellipodia of Cos-7cells [120], suggesting that contact is
maintained during the movements of these organelles. This
shows that some contact sites can be pretty stable. However
it is not the rule. Many interorganelle structures display very dynamic behaviors, such as the STIM–ORAI interaction (1.1.2.2),
or the ORP1L-mediated ER–endosome contact [94,95]. STIM
and ORAI only interact upon ER-Ca 2+ depletion and ORP1L is
recruited to endosomes upon endosome sterol repletion. What
is less clear is whether these dynamic proteins are recruited to
static contact sites, or whether these sites are newly formed
by the recruitment of these proteins. When SOCE is stimulated
repeatedly, STIM can be recruited to the same locations, suggesting that these locations could be contact sites that are
maintained between SOCE stimulations [37]. On the other hand,
the association between the ER and PM, as detected by transmission electron microscopy, is increased upon SOCE, or by
STIM overexpression, suggesting that STIM also has the ability
to induce tethering [40]. This tethering could operate by
extending preexisting contact sites, or by creating contact sites
de novo. The nucleus–vacuole junction (NVJ) also appears to be
a dynamic structure. Yeast cells in exponential phase do not
have much of a NVJ, but entry into stationary phase, nitrogen
limitation or rapamycin treatment triggers the expansion of NVJs
[116]. The mechanisms underlying this induction are unclear, and
could include transcriptional induction of the proteins involved in
tethering. Indeed, the tethering protein Nvj1 is induced by a variety of stresses dealing with starvation response, including entry into stationary phase [158], and its artificial overexpression alone
can induce the formation and spreading of NVJs [115]. Yet transcriptional induction of Nvj1 is likely not the only player, as cells
constitutively overexpressing Nvj1 are still able to respond to starvation by making even more NVJs [117].
1.1.3. The ER–mitochondrial Ca 2+ crosstalk
1.1.3.1. The role of contact sites. Back in the 1960s, isolated mitochondria were shown to take up Ca 2+ when externally applied in vitro (for
an historical review, see [44]). This import is dependent on a mitochondrial Ca 2+ uniport (MCU) [45,46] on the inner mitochondrial
membrane (IMM) and on porins, which are non-selective OMM
pores for small molecules. This phenomenon also requires an intact
membrane potential across the IMM. However, the physiological relevance of these findings was soon questioned as the [Ca 2+] necessary
to activate the uniport were orders of magnitude higher than the
physiological [Ca 2+] found in the cytosol. The conclusion at the
time was that mitochondrial [Ca 2+] import should never take place
in a healthy cell. This view changed with the advent of methods
allowing in vivo organellar [Ca 2+] monitoring, which showed that mitochondrial [Ca 2+] increased dramatically upon opening of the IP3R
in live cells [47]. Yet, in the same conditions, the cytosolic [Ca 2+]
did not increase to sufficient levels to activate the MCU, raising a paradox. This paradox was resolved by the following hypothesis: due to
the slow diffusion of Ca 2 +, the local [Ca 2+] around the mouth of
the IP3R may be much higher than in the rest of the cytosol. A close
proximity between the ER and the mitochondria may thus allow activation of the MCU [48]. This hypothesis established the framework
for an abundance of studies aimed at revealing the physiological
importance of ER–mitochondria connections in Ca 2+ signaling.
Measuring [Ca 2+] at ER–mitochondria contact sites using genetically
encoded [Ca 2+] reporters artificially targeted to these sites (see
Box 3) showed that they indeed represent microdomains of the cytosol, where the [Ca 2+] can raise to the micromolar range, compatible
with the activation of the MCU [49]. Moreover, expressing artificial
ER–mitochondria tethers made of a protein that bears both ER and
mitochondrial targeting sequences (see Box 3), can increase the coupling between the ER-efflux and mitochondria-influx, again underlining the importance of ER–mitochondria MCSs in Ca 2+ exchange
[49–51].
Thus, mitochondria take up Ca 2+ owing to ER–mitochondria
MCSs. What is the physiological function of this uptake? Three roles
have been proposed. First, Ca 2+-dependent enzymes are found in
the mitochondrial matrix and may require mitochondrial Ca 2+
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
4
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
uptake for their proper function. Second, as Ca 2+ is toxic in the cytosol, mitochondria could act as Ca 2+ buffers to limit the cytosolic Ca 2+
increase following IP3R stimulation. Third, mitochondrial Ca 2+ could
act as an important trigger for mitochondrial apoptosis.
1.1.3.2. Ca 2+-dependent respiration. Pyruvate dehydrogenase, NAD+isocitrate dehydrogenase and 2-oxoglutarate dehydrogenase are
three mitochondrial enzymes centrally located in the Krebs cycle that
are activated by Ca 2+ [52]. Other Ca2+-regulated mitochondrial activities include ATP/ADP exchange, ATP/Pi exchange and ADP phosphorylation by the F0/F1 ATPase [53]. Thus, mitochondrial energy production
is coupled to mitochondrial Ca2+ uptake. How important is that for
cellular bioenergetics? This question should optimally be addressed
by directly manipulating the players of mitochondrial Ca2+ influx,
which was not possible until the recent discovery of MCU [45,46].
Interesting leads, nonetheless, came from manipulating a process upstream of mitochondria Ca2+ influx, i.e. ER efflux. When knocked-out
of all isoforms of IP3R, chicken cells react by increasing autophagy
[54]. Autophagy is a cellular mechanism usually induced in response
to nutrient limitations, suggesting that cells devoid of IP3R are
experiencing starvation. The reduced or abrogated ER–mitochondrial
Ca2+ exchange in these cells could thus lead to an under-stimulation
of the above-mentioned enzymes, leading to decreased ATP production, to which cells respond by mounting a starvation response. In
support of this model, the AMPK pathway is activated in these cells,
indicating a low ATP/ADP ratio [54].
1.1.3.3. Cytosolic Ca 2+ buffering. During IP3R or RyR activation, all the
Ca 2+ that ends up in mitochondria does not end up in the cytosol.
This could imply that mitochondria have a buffering role by avoiding
that excess Ca 2 + reaches the cytosol. Stimulating IP3R or RyR in
conditions that inhibit mitochondrial Ca 2 + uptake (for example in
presence of a mitochondrial uncoupler that dissipates mitochondrial
membrane potential) results in a much bigger cytosolic [Ca 2+] increase [55,56]. As cytosolic Ca 2+ is a potent trigger for necrosis, this
buffering may have cytoprotective effects.
1.1.3.4. Induction of apoptosis. Beside its potential cytoprotective action, mitochondrial Ca 2+ may also have a proapoptotic role. Isolated
mitochondria treated with high [Ca 2+] in vitro undergo opening of
the so-called permeability transition pore (PTP). Although original
studies describe the PTP as a pore opening in both mitochondrial
membranes independent of MCU activity [57], the current consensus
is that PTP is a pore in the OMM, which is opened by Ca 2+ overload in
the matrix (for review see [58]). Opening of this pore causes the release of small inter-membrane space proteins, such as Cytochrome
C, which triggers a cascade of events leading to programmed cell
death.
Could ER–mitochondria Ca2+ exchange similarly lead to PTP opening? Several lines of evidence suggest that this is the case. Antiapoptotic
proteins such as Bcl-2 were shown to lower ER [Ca2+] when overexpressed, which in this context, could explain at least part of their
antiapoptotic function [59]. Similarly, deletion of proapoptotic proteins
led to decreased ER [Ca2+], in turn leading to decreased apoptotic
sensitivity [60]. More directly involving Ca2+ homeostasis, a mutation
in the ER Ca2+ pump SERCA1 was shown to act as an oncogene by
inhibiting apoptosis, suggesting again that high ER [Ca2+] is necessary
to activate the PTP in the mitochondria [61]. Similarly, downregulating
IP3R renders cells resistant to a variety of apoptotic stimuli [62,63]. Thus,
direct ER–mitochondria Ca 2+ exchange is a likely trigger of the mitochondrial pathway of apoptosis.
The experiments described in Sections 1.1.3.2, 1.1.3.3 and 1.1.3.4
assessed the importance of mitochondrial Ca2+ influx by manipulating
ER efflux or mitochondrial membrane potential, which are bound to affect more than just mitochondrial Ca 2+ influx. The recent identification
of the molecular players of mitochondrial Ca2+ influx [45,46] will bring
the power of genetics to assess the physiological importance of this
phenomenon. It is notable that mice could be treated with MCU RNAi
for weeks without noticeable effects on their health. Moreover, mitochondria isolated from these animals showed normal bioenergetics
parameter despite their near complete loss in Ca 2+ uptake [45]. Closer
inspection will be required to clarify the role of mitochondrial Ca2+
uptake in the above processes, and indeed the tools necessary for this
examination are now available.
1.2. Interorganelle lipid exchange
Cellular lipids fulfill a wide range of functions. They can act as
energy storage in the form of triglycerides, as important parts of
intra- and intercellular signaling cascades and as main building blocks
of bio-membranes. Nowadays, literature describes more than 1000
different lipid species in eukaryotic cells [64], distributed asymmetrically between organelles and even between the two leaflets of one bilayer [65]. Phosphatidylcholine (PC), for instance, is not only the most
common lipid in the cell but also most abundant in the endoplasmic
reticulum (ER). Phosphatidylethanolamine (PE) and cardiolipin
(CL), in contrast, are significantly enriched in mitochondria. CL
concentrations are furthermore up to four times higher in the IMM
compared to the OMM [66,67]. These asymmetric distributions have
functional consequences. For instance, the compact packing of
sphingolipids and sterols in the plasma-membrane, where these
lipids are enriched allows this membrane to resist increased levels
of mechanical stress [66]. The asymmetric distribution of cellular
lipids is partly explained by their asymmetric synthesis and breakdown. For instance a complex network of phosphatidylinositol (PI)
kinases and phosphatases establishes and maintains a gradient of different phosphoinositides on the different cellular membranes. However, most lipids are synthesized in one place – the ER – suggesting
the existence of an active and regulated lipid sorting system. The vesicular transport system shuttles lipids together with proteins across
the endomembrane, but this mode of transport is far from the whole
story. PE, cholesterol and ceramide can for instance reach the GOLGI
and PM even when vesicular transport is inhibited [68–70]. More importantly, mitochondria are completely disconnected from vesicular
transport routes although they are centrally involved in cellular
lipid homeostasis. Indeed, the de novo aminoglycerophospholipid biosynthesis pathway is spatially segregated between ER and mitochondria. Phosphatidylserine (PS) is made in the ER membrane by the PS
synthase (PSS). PS is then decarboxylated to PE by a decarboxylase
(PSD) that is integral to the IMM. PE is further processed to PC by
three sequential methylation steps performed by ER-resident proteins (Fig. 2D). This convoluted biosynthesis pathway involves a
back-and-forth journey from the ER to the IMM. Moreover, PC,
which is abundant in both mitochondrial membranes, needs to travel
back to the mitochondria (for review, see [71]).
In this context, MCSs between pairs of organelles have long been
thought to facilitate lipid exchange by ensuring the specificity and
efficacy of the exchange reaction [72].
1.2.1. Lipid transport proteins
Non-vesicular lipid transport is catalyzed by lipid transport proteins (LTPs), which are able to extract one lipid molecule from a
membrane and deliver it to another. This type of transport involves
special lipid-transport domains (LTD) that can shelter the hydrophobic moieties of lipids from the aqueous environment (Fig. 2A and B).
The mode of action of these proteins has been extensively studied
in vitro and corroborated by structural analyses (for reviews see
[73,74]). Different types of LTD bind different lipids. In addition to
their LTDs, LTPs often contain different combinations of targeting
domains (Fig. 2A), which direct them to special compartments.
For example, the diphenylalanine-in-an-acidic-tract (FFAT) motif
binds to the ER protein VAP (VAMP-associated-protein) [75–77], the
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
5
A
B
C
D
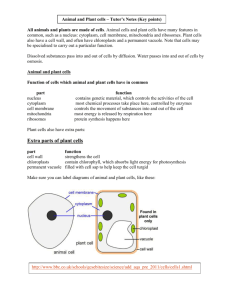
Fig. 2. MCSs and lipid homeostasis. A) Non-exhaustive list of LTPs and their domain organization. Many of these LTPs are found at MCSs. ORPs bind to sterols and to PI4P. The steroidogenic acute regulatory protein-related lipid transfer (START) domain can bind either sterols, phospholipids or ceramides. PITP and Sec14-like domains bind PC and PE.
Lipid-binding by SMP-containing proteins is not experimentally established, but SMP is remotely homologous to the TULIP domain. B) LTPs often show affinity for two or more
lipid species. Here are shown: PITPα bound to a PC and a PI molecule, Sph1 (a Sec14-paralog) bound to a PI and PC molecule. CERT (START-containing) bound to a ceramide
and a DAG molecule and Kes1 (ORP) bound to an oxysterol and a PI4P molecule. While the polar heads can adopt different conformations (see Sph1 and Kes1), the hydrophobic
tails bind in the same pocket, showing that only one molecule can be transported at a time. The crystal structures used in this figure have the following pdb IDs: 1t27 and 1uw5
for PITPα, 3b7q and 3b7N for Sph1, 2e3q and 2z9y for CERT and 3spw and 1zhw for Kes1. C) Model of how a gradient in sterol and PC is generated by the interconnected cycle
of ORPs (blue) and PITP/Sec14 proteins (yellow). ORPs exchange sterols for PIPs. PIPs being abundant in the late secretory organelles and rare in the ER, due to the action of
PI4P phosphatases (red), this creates a net transport of sterols toward the late-secretory organelles. PITP/Sec14-like proteins exchange PI for PC. PI being actively removed from
the Golgi by phosphorylation by PI kinase (green), the PITP exchange reaction creates a net transport of PC from the Golgi to the ER. Combining these two exchange reactions results
in a net PC-sterol exchange that is fueled by ATP hydrolysis by the Golgi PI-kinase (green). D) de novo PC biosynthesis requires two ER–mitochondria exchange reactions.
Phosphatidylserine (PS) is synthesized in the ER. It is converted to phosphatidylethanolamine (PE) in the IMM by the PS-decarboxylase (Psd). PE is further converted to phosphatidylcholine (PC) by three methylation steps, which take place in the ER.
Pleckstrin-Homology (PH) domains bind to specific PIPs found in the
Golgi and the plasma membrane [78–81], and the ArfGAP1-LipidPacking-Sensor (ALPS-) motifs, which bind to highly curved membranes [82]. LTPs bearing two targeting motifs for two different membranes are thus naturally directed to MCSs between these two
membranes. One intriguing feature of many LTDs is their ability to
bind two lipid species in a competitive manner. Many LTPs have
been crystallized in complex with different lipids. This is the case of
the PI Transporter Protein (PITP) with PI and PC [83,84], the ceramide
transporter CERT with ceramide and diacylglycerol (DAG) [85], and
the Oxysterol-binding-protein-Related-Domain (ORD) of Kes1 with
sterols and PI4P [86,87] (Fig. 2B). In these examples, whereas the
polar head of lipids contact the protein at sites that are specific for
each lipid species, the aliphatic tails occupy the same hydrophobic
pocket. This renders the binding of two lipids mutually exclusive.
This feature suggests that many of these proteins function as lipid
exchanger rather than unidirectional transporter.
1.2.2. Non-vesicular lipid exchange in lipid homeostasis
Current models suggest that specific lipid exchange reactions
may happen at MCSs. For example, a large subfamily of LTPs has an
Oxysterol-binding-protein-Related-Domain (ORD) domain, and is
therefore called ORP (oxysterol-binding protein related proteins).
The ORD domain is highly conserved and binds a variety of sterols.
It has recently been found to also bind PI4P competitively with sterols
[87], and to promote the trading of one lipid molecule against the
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
6
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
Box 2
Contact-sites and lipid rafts.
Lipid rafts are thought to be assemblages of lipids and proteins that are laterally segregated from the rest of the membrane through a combination of protein–protein and protein–lipid interactions, lipid phase immiscibility and association with the cytoskeleton [159]. They have
long been operationally defined as detergent-resistant membranes (DRM). Indeed, they resist extraction with 1% Triton X100 at 4 °C, a
property that has been conveniently used for their purification and analysis [160]. However, due to their small size, lipid rafts have never
been directly observed in live cells and indirect methods are used to infer their existence, which has thus long remained controversial
[161]. Rafts are enriched in select proteins and in lipids, such as sterols and sphingolipids. Several lines of evidence make a connection between membrane contact sites and lipid rafts. Mitochondria-associated membranes (MAMs) are operationally defined as an ER fraction that
cosediments with mitochondria during subcellular fractionation and are therefore thought to represent the subfraction of the ER that is attached to mitochondria in vivo (see Box 3). The MAM fraction has characteristics of lipid rafts. First, several analyses show that the MAM
fraction is detergent resistant [162,163]. Second, this fraction is enriched with sterols and simple sphingolipids [162,164,165]. Many
MAM-enriched proteins have been shown to associate with DRMs, including the Sigma-1 receptor, the presenilin 1 and 2, PEMT, Vdac1
and mitofusin-2 [162,163]. ER–PM contact sites may also have raft characteristics. When STIM is recruited to ER–PM contact sites, it concomitantly relocates to DRMs, suggesting that ER–PM contact sites are lipid rafts [166]. Finally, the nuclear vacuole junction (NVJ) shares
common points with rafts, as select vacuolar proteins are enriched or excluded from NVJs [117]. While enrichment of a protein at NVJ can
be explained by direct interactions of these proteins with the NVJ structural components Vac8 and Nvj1, selective exclusion of vacuolar
proteins is more difficult to explain. One model is that the lipids in the vacuole may also be laterally segregated, leading to enrichment
and exclusion of select proteins that have differential affinities for different lipid phases, akin to canonical lipid rafts. In support of this notion, pharmacological or genetic inhibition of very long-chain fatty acid synthesis affects NVJ morphology and function, suggesting a role
for specific lipids in the assembly of these contact sites [119].
It is surprising to find ER proteins on lipid rafts. The lipid phase-separation required to assemble rafts is supposed to take place only on membranes with sufficient sterols and sphingolipids, and therefore the sterol- and sphingolipid-poor ER membrane should be devoid of canonical
rafts [66]. Yet, since these DRM-associated ER proteins are found at contact-sites, it may be that the ER membrane in which they insert is
indeed detergent soluble, but that the membrane to which they associate in trans is a DRM. For example, activated STIM interacts with
negatively-charged lipids on the plasma membrane through a polybasic domain on its cytosolic domain. Therefore, even if the ER membrane
is totally soluble, STIM may remain associated with detergent-resistant bits of the PM. This explanation cannot be invoked for MAMs, as
both ER and OMM are supposed to be devoid of canonical lipid rafts. As MAM-derived DRMs contain both ER- and OMM-integral proteins, it
is not clear which membrane is detergent-resistant. It is notable, however that drugs that disturb rafts by affecting sterol concentration
(such as methyl-beta-cyclodextrin, which sequesters sterols out of membranes), cause relocalization of MAM proteins. This relocalization
can be observed both at the level of the biochemical MAM fractionation and at the level of the subcellular localization [162].
other in an in vitro assay. Therefore, although the in vivo relevance of
their LTP activity is questioned [88], ORPs have been proposed to
transport sterols from the ER to the Golgi and PM. Indeed, many
ORPs localize at MCSs between these compartments (see Section
1.2.3). Because PI4P is immediately dephosphorylated in the ER by
specific phosphatases [89], this exchange reaction should only go
one way. Sec14 on the other hand can transport both PC and PI between the ER and the Golgi [90]. In this model, the ATP dependent
conversion of PI to PI4P by Golgi resident PI kinases provides the
directionality for the reaction. Thus, coupling both cycles of ORPs
and Sec14 could provide a neat device that uses the power of ATP to
pump sterol into the late secretory pathway (Fig. 2C, [87]).
The genetic dissection of these pathways is complicated by the
pleiotropic roles of lipids in membrane biology. PI4P for instance is a
central regulator of vesicle trafficking. Another lipid transporter
tightly linked to vesicular traffic is CERT (Fig. 2A–B), which transports
ceramide from the ER to the Golgi [75], where it is used for the production of DAG and sphingomyelin [91]. DAG is important for vesicle
budding from the trans-Golgi [92], showing again the entanglement
of lipid transport with other aspects of membrane biology.
1.2.3. MCSs in lipid exchange
While energy-dependent processes provide directionality to lipid
exchange, MCSs may provide specificity. Many LTPs are targeted to
MCSs. For example, both CERT and several ORPs have two targeting domains: one PH-domain and one FFAT motif. The FFAT-motif anchors
these LTPs to the ER by interacting with the ER-protein VAP, while
the PH domain binds to PIPs on closely apposed membranes [74]. For
instance, the yeast ORP Osh1 is found at a contact site between the
perinuclear ER (the nuclear envelope), where it interacts with yeast
VAPs (Scs2 and Scs22 [76]), and the vacuole (the yeast lysosome),
which it contacts via its PH domain [93] (Fig. 3). This Nuclear-ER
Vacuole Junction (NVJ) is a contact site formed dynamically with
a function that is only partly understood (see Section 1.3.1). The mammalian ortholog Oxysterol-binding-protein-related-protein 1L (ORP1L)
is also involved in dynamic contact sites between the ER and lateendosomes/lysosomes. In this case, ER-endo-/lysosomes contact sites
are formed in response to low cholesterol, which is sensed by ORP1L's
ORD. Formation of the ER/endosome contact site by binding of ORP1L's
FFAT domain to VAP causes dissociation of dynein-binding proteins
from the late endosome, resulting in a cellular redistribution of these
endosomes [94–96]. Thus LTPs cannot only exchange and transport
specific lipids across the aqueous phase of the cell, but they can also establish specific contact sites and serve as sensors of lipid levels between
two compartments.
As stated before, ORD domains are found in many eukaryote proteins. For example, yeast has seven ORPs. Four of them (Osh2, Osh3,
Osh6 and Osh7) localize to contact sites between the ER and the
plasma-membrane [76,89,97]. Two of these four (Osh2 and Osh3)
harbor a FFAT and a PH domain necessary for their localization
(Fig. 2A). The other two are targeted to contact sites by unknown
mechanisms [98]. Moreover, sterol–PI4P exchange activity appears
to be one of the most conserved features of ORDs [87].
1.2.4. Lipid exchange at ER–mitochondria MCSs and the SMP domain
Although the prime example for MCS-dependent non-vesicular
lipid exchange is the ER–mitochondria connection (see Section 1.2),
it is unsettling to note that the LTPs involved are mostly unknown.
In yeast, the best candidates are the members of the ER–Mitochondria
Encounter Structure (ERMES). This protein complex is made of
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
7
Fig. 3. Established and potential tethers at various MCSs. 1) Potential tethers between the ER and the PM in metazoan, yeast or both. For explanations on individual proteins and
complexes, see Section 2.3. 2) Potential tethers between the ER and the mitochondria. See Section 2.2. 3) Potential tethers between the ER and the vacuole at the nucleus–vacuole
junction (NVJ) in yeast. The interaction of Vac8 and Nvj1 constitutes the main tether (see Section 2.1), but Osh1 interaction with Scs2 and Scs22 may also participate in tethering.
4–6) Potential tethers between the ER and the late endosome, the Golgi apparatus and lipid droplets. See Section 2.4.
five subunits. One of the subunits (Mmm1) is integral to the ER
membrane, while two others (Mdm10 and Mdm34) are inserted in
the OMM [99–101]. This complex was identified based on its ER–
mitochondrial tethering activity: its function can be bypassed by the
expression of an artificial ER–mitochondria tether (see Section 2.2.1
and Box 3). Deletion of any member of the ERMES complex causes
pleiotropic phenotypes that are difficult to decipher [102]. Yet, interesting insight came from an unbiased genetic interaction screen that
detected a strong correlation in the patterns of genetic interaction
of ERMES mutants and that of strains lacking the mitochondrial PS
decarboxylase (Psd1, Fig. 2D). That Psd1 substrate and product have
to respectively come from and return to the ER (Fig. 2D), suggests a
role for ERMES in lipid transport. Whether ERMES may itself act as
an LTP for this transport reaction is a matter of debate. On the one
hand, recent bioinformatic analyses suggested that ERMES proteins
may directly bind lipids. Three ERMES components (the ER protein
Mmm1, the cytosolic Mdm12 and the OMM protein Mdm34) share
a protein domain, called SMP, which is common to a group of conserved eukaryotic Synaptotagmin-like, Mitochondrial and PH-domain
containing proteins (SMP, Fig. 2A) [103,104]. Homologies between
these proteins are difficult to see at the level of the primary sequence,
but are more obvious using hidden Markov models that take the predicted secondary structure into account. Additional homology prediction shows that the SMP domain is part of a larger protein family with
structurally characterized members. Members of this family harbor
an interesting structure made of a very elongated hydrophobic
groove, which binds various lipids and hydrophobic molecules. For
this reason, the family was named TULIP (for tubular-lipid-binding)
[104]. Phospholipids can be bound by TULIP proteins. Interestingly,
in this case, the hydrophilic heads “snorkel” out of the groove and
do not make any contact with the protein. Thus, phospholipid binding
by TULIP proteins appears more or less independent of the lipid species. Whether SMP domains are also able to bind lipids in a hydrophobic groove is unclear, but all other yeast SMP-containing proteins
(Nvj2, Tcb1, Tcb2 and Tcb3) were also found to localize at various
MCSs [105]. SMP must therefore have a common function at MCSs,
and it is tempting to speculate that it is lipid exchange.
But on the other hand, some data conflict with the idea that
ERMES members act as LTPs. First, ERMES deletion only has a limited
impact on ER–mitochondria lipid exchange [99,106] – whether it has
any effect at all is even controversial [107] – showing that ERMESindependent lipid exchange can take place. Second, ERMES was identified as a tether because its absence could be compensated for by
an artificial ER–mitochondria tether. Two of the three SMP-bearing
ERMES components Mdm12 and Mdm34 are dispensable as long
as this artificial tether is present. Since this artificial tether is not
expected to work as an LTP, it seems that the LTP activity of Mdm12
and Mdm34, if any, is not required under laboratory conditions [99].
In mammals, where ERMES is not conserved, the identity of ER–
mitochondria LTPs is even more mysterious.
1.2.5. Lipid storage
Finally, one important function of lipids is their use as energy storage. This storage takes place mainly in the form of neutral lipids, such
as triglycerides stored into intracellular lipid droplets. Lipid droplets
(LDs) are thought to emanate from the ER by triglyceride and steryl
esters accumulation in between the two leaflets of the ER membrane
(reviewed in [108] and [109]). As a result, LDs have a core of neutral
lipids surrounded by a single leaflet of amphipathic lipids. It is
unclear, whether continuity between the ER membrane and the single leaflet surrounding LDs is maintained during the whole lifetime
of the LDs [110]. It is clear, however, that LDs are covered with closely
apposed ER cisternae on most of their surface [111–113]. These ER–
LD contact sites likely serve in translocating newly synthesized neutral lipids from the ER to the core of the LDs [114].
1.3. Other functions of MCSs
Besides lipid and Ca 2+ exchange, a few example MCSs perform
additional functions in organelle biology.
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
8
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
Box 3
Methodologies to study contact sites.
The complete toolbox of modern molecular and systems biology is available for the study of MCS, but a few methods have proven particularly useful for this purpose and will be discussed here.
• Biochemical purification of MCSs. Classical subcellular fractionation can separate different organelles according to their densities or
sedimentation coefficient using isodensity or velocity centrifugation, respectively. But could organelles that are physically tethered
have special physical characteristics that allow purification? This is the rationale behind purification of Mitochondria-associated
membranes (MAM) [167]. Purification of mitochondria commonly involves a step of differential velocity centrifugation during
which mitochondria sediment at a lower speed than the bulk ER. However, following this step, mitochondria are found heavily contaminated with ER membranes. Thus, mitochondria have to be separated from the contaminating ER by a step of isodensity centrifugation across a continuous or step gradient of sucrose, Percoll, Optiprep™ or Nycodenz®. The ER contaminant that is retrieved
from the gradient has a different composition from that of bulk ER. This ER fraction, originally dubbed “fraction X” [72] is therefore
thought to represent a sub-domain of the ER physically attached to mitochondria. This strategy was emulated in the search for
other fractions such as PM-associated membranes (PAM) [168,169] or plastid associated membranes (PLAM) in plants and
algae [170]. The MAM fraction is enriched for many lipid-related enzymes and Calcium signaling proteins, consistent with a role
of ER–mitochondria connection in these two processes (for review see [171]). Yet the definition of MAM as the “fraction of the
ER that cosediment with mitochondria” is an operational definition, just as “detergent-resistant membranes” is an operational definition for lipid rafts (Box 2). The nature of the ER found in the MAM fraction is poorly defined. Is it only the lipid and proteins directly
attached to mitochondria? Is it the mitochondria-attached ER plus just enough ER to seal a vesicle around it? Is it an undetermined
amount of ER that stays connected to the mitochondria-attached ER? Moreover, as any subcellular fraction, MAM is not completely
pure. Indeed, most proteins retrieved in the MAM fraction are not ER proteins [172]. Other methods, such as microscopy, aimed at
localizing MAM-associated factors with respect to ER and mitochondria do not necessarily clarify the picture, as the subcellular localization of MAM-enriched proteins is not evident. The canonical MAM protein FACL4 is found at the whole mitochondrial surface
[173]. The same is true for AMFR/GP78 [174], or the presenilins 1 and 2 [173]. By contrast, other highly MAM-enriched proteins
such as the Sigma-1 receptor [175] or Ero1alpha [176], show a staining that overlaps only with select regions of mitochondria,
with most of the signal nowhere close to them. To complicate things, different studies disagree on the extent of MAM enrichment
of different factors. For example, the amount of IP3R3 found in the MAM differs between studies [143,144]. Even in a single study,
when virtually all IP3R3 is found in the MAM by subcellular fractionation, only a fraction of it is found close to mitochondria by immunofluorescence [144]. Thus, akin to lipid rafts ten years ago, much needs to be done to take MAMs from an operational definition to a solidly-grounded biological reality.
• Microscopy is of course a method of choice to visualize the encounter of two organelles. Organelle-targeted fluorescent proteins
are perfect markers for live-cell microscopy. However, contact sites per se are happening at distances (typically less than 30 nm)
far below the resolution limit of light microscopy, making it difficult to discriminate genuine contact sites from mere colocalization.
For instance, a recent study shows that a cell line that displays reduced ER–mitochondria colocalization as assessed by confocal
light microscopy, actually shows increased ER–mitochondria contacts by electron microscopy [177]. Thus, care must be taken
when analyzing colocalization of organelles at light-microscopy resolution, as changes in colocalization may reflect changes in organelles distribution rather than changes in connection. These limitations can be circumvented by choosing proper imaging techniques. For example, TIRF microscopy is a method of choice to visualize ER–PM connections in adherent cells (for examples see
[12,19,25,28]). Since the evanescent field of total internal reflection is narrow, the ER only enters this field when in a range
of 100 nm from the cover-slip-bound PM. Although the 100 nm-wide evanescent field of TIRF microscopy is still quite larger
than the range of interorganelle connection, TIRF performs much better than confocal microscopy that displays a resolution in
Z in the order of the micrometer. Another way to circumvent resolution issues is to image areas of the cells where organelles
are sufficiently scarce to be well resolved. The lamellipodia of Cos-7 cells, for instance, are very thin (typically 100–200 nm
thick) [178], abrogating the problem of Z-resolution. The confinement of the cytoplasm in these lamellipodia has two additional effects: First it causes the spreading of the cellular components, which allows resolution of individual organelles such as
mitochondria, ER cisternae and ER tubules [120]. Second, it forces intersecting organelles against each other (mitochondria,
for instance, are typically 300–500 nm thick and must thus flatten to fit into lamellipodia [179]). As a consequence, every
observed organelle intersection represents a contact site de facto [134]. Electron microscopy does not suffer from resolution
limitations, at the expense of versatility and the possibility to study live cells. New light microscopy techniques that are not
limited by Abbe's law, approach the resolution needed to study contact sites and will likely provide invaluable tools for this
purpose.
• Genetic screens have proven extremely useful to discover molecular players of organelle contact sites. RNAi screens have been
at the root of the discovery of the components of SOCE [12–16]. In the cases of both STIM and ORAI, screens were designed
to identify RNAi targets that affect SOCE. Identification of these factors later led to the discovery of the contact-sites where
ER–PM communication was taking place. In this case, the function led to the structure. A difficulty in analyzing the results
of such screens is that they are prone to hit functionally quite far from their targets. And much work may remain to bridge
the function to the structure. For example, an RNAi screen has identified LETM1 as a protein necessary for Ca 2 + import into
mitochondria [180]. Yet, other studies indicate that LETM1 acts as a K +/H + exchanger [181,182]. It is thus still controversial
whether a defect in K +/H + exchange causes the Ca 2 +-import defect or if LETM1 can also transport Ca 2 +. But screens can
also be designed to seek for mutants with reduced or abolished tethering activity. The tethering activity of the ERMES complex
was discovered following this principle [99]. Mutant yeast cells were selected based on their ability to be complemented by an
artificial ER–mitochondria tether (see below). Such mutants were part of a single complex found at the interface of the ER and
the mitochondria, which is involved in a plethora of processes dealing with virtually all aspects of mitochondrial biology. Thus
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
9
Box 3 (continued)
in this case, the structure leads to the function. The drawback of this reversed approach is that the real function of these contact sites remains not fully understood [102,107].
• Artificial tethers represent a powerful way to approach MCSs. Pioneered by Hajnóczky and colleagues, this approach consists in
encoding targeting signals for different organelles into the same protein, or into different proteins that will later be joined together
through dimerization motifs [49,51]. Such synthetic proteins can act as artificial tethers, thereby increasing the connection between
two organelles. This feature was used to show that increased ER–mitochondria connection caused increased Ca 2+ exchange between both organelles [51], and also to find mutants of natural tethers that relied on an artificial tether for growth (see above) [99].
Additionally, due to their dual affinity, such synthetic proteins can bring reporters to defined contact sites. For instance, artificial
tethers with fluorescent moieties allow localization of contact sites within the cell. Similarly, artificial tethers fused to Ca 2+ biosensors
showed that local [Ca 2+] at ER–mitochondria connections were ~25-fold higher than in the rest of the cytosol [49]. However these
tethers have the potential to generate non-physiological situations. Indeed, it is virtually impossible to target an artificial protein to a
contact site using two targeting sequences without concomitantly increasing the tethering forces between both organelles involved.
For instance, Csordás et al. designed a FRET-based reporter that targets the ER–mitochondria interface thanks to a rapamycin-induced
dimerization between the ER-targeted FRET donor and the mitochondria-targeted acceptor. Upon rapamycin treatment, FRET signal,
which is originally found at few discrete foci, progressively expands to cover almost all the mitochondrial surface, due to the intrinsic
tethering activity of the reporter [49]. Care should thus be taken when using such synthetic proteins.
1.3.1. Piecemeal microautophagy of the nucleus
In yeast, under nutrient limiting conditions, the vacuole becomes
the largest cellular organelle and is used for the autophagic recycling
of parts of the cytoplasm. With these conditions, a MCS between the
nuclear ER and the vacuole (nuclear–vacuole junction, NVJ) [115]
becomes the site of a special type of autophagy, whereby bits of the nucleus are delivered to the vacuole. The NVJ grows during starvation to a
point where it covers a significant fraction of the nucleus. This MCS
then invaginates into the vacuole. At the final stage, scission of the ER
and fusion of the vacuolar membrane release a vesicle containing
nuclear cargo into the vacuole lumen, where degradation occurs.
This phenomenon is therefore called piecemeal microautophagy
of the nucleus (PMN) [116]. Many aspects of PMN remain unclear.
(1) While the machineries that are responsible for the formation of
the NVJ are well-characterized (see Section 2.1), those involved in
the scission of the ER and abscission of the vacuolar vesicle are unknown. (2) The lipids in the interior face of the vacuole are resistant
to degradation by vacuolar lipases, which is important to maintain
the integrity of the vacuolar membrane. It is thus not clear how the
vesicle, once detached from the vacuolar membrane, becomes a substrate for degradation while the same membrane, when still connected
to the vacuolar membrane is not. This distinction could be related to a
diffusion barrier established at the NVJ, which may exclude select lipids
and proteins from the forming vesicle [117] (Box 2). Alternatively, recruitment of lipid modifying enzymes to the NVJ, and thereby to the
PMN-vesicle membrane, could locally change the lipid composition of
this membrane [118,119], rendering it selectively sensitive to the vacuolar lipases. (3) The function of PMN is unclear, especially since mutations disabling this process do not yield notable growth defects [116],
but roles can be speculated. For instance PMN could regulate the size
of NVJ by deleting overgrown interfaces. Alternatively, since the nucleolus is usually the nuclear region adjacent to the NVJ, PMN could serve
in the recycling of ribosome biogenesis factories [116].
1.3.2. Mitochondrial dynamics
Mitochondria are highly dynamic organelles. They represent, after
the ER, the second largest intracellular membrane network. This network is constantly remodeled by fusion and fission events, and by cytoskeletal driven movements that allow faithful inheritance during
asymmetric cell division and allow mitochondria to reach distant
places in the cell, such as the neuronal synapse. ER–mitochondria
contact sites have recently been shown to play a role in all of these
aspects of mitochondrial dynamics.
1.3.2.1. Mitochondrial movement. Pioneering work in the lab of Gia
Voeltz showed that ER and mitochondrial dynamics are coupled
[120]. By imaging very flat lamellipodia, Voeltz and colleagues could
resolve individual mitochondria and individual ER tubules. Both organelles move in a coordinated manner on a subset of acetylated microtubules. Mitochondrial movement along microtubules is a well-studied
phenomenon and one of the best-characterized adapter proteins in
this process is the GTPase Miro [121]. Miro is a mitochondriatail-anchored protein that contains two GTPase and two Ca2+-binding
domains. Miro, anchored to the OMM binds to kinesin heavy-chain via
a second adaptor protein. This binding is necessary for mitochondrial
movement along neuronal axons. But the surprising feature of Miro is
its ability to stop mitochondrial movement upon high [Ca 2+] [122].
This phenomenon is dependent on both Miro's Ca2+-binding domains
and occurs by sequestration of kinesin heavy chain motor head away
from microtubules. One of the two mammalian paralogs of Miro,
Miro-1, is found at ER–mitochondria interfaces in the lamellipodia of
Cos-7cells, linking mitochondrial movement and ER–mitochondria
connection [100]. Moreover, Gem1, the unique yeast Miro, is a member
of the ERMES complex [100,101]. These findings may partly explain the
coupling between ER and mitochondrial dynamics. The proximity of
the ER may provide the Ca2+ necessary to regulate Miro. In support
of this notion, expression of a mutant form of VAPB, involved in
type-8 amyotrophic lateral sclerosis (see Section 2.2.4), increases
ER–mitochondria Ca 2+ coupling and inhibits mitochondrial movement
in a way that depends on Miro Ca 2+-binding domains [123]. The
functional role of the coupling of ER and mitochondrial movement is,
however, unclear.
1.3.2.2. Mitochondrial fusion and fission. Both mitochondria fusion and
fission involve GTPases [124–127]. During fission, the dynaminrelated protein 1 (Drp1) assembles into helical oligomers that circumscribe the mitochondrial tubule. GTP hydrolysis then provides
the energy to squeeze the mitochondrion down to a point of fission
[125,126,128–130]. In fusion, Mitofusins (MFNs) on opposite mitochondria interact with each other while GTP hydrolysis forces the
fusion of the two membranes [127,131]. In mammals, two MFNs
(Mfn1 and Mfn2) are necessary for this function, therefore deletions
of either or both result in fragmented mitochondria [131]. Mfn2
may have acquired an additional function besides its role in mitochondrial fusion, namely, ER–mitochondria tethering. Mfn2 KO cells
have a fragmented ER, which can be rescued by expressing an
ER-targeted version of Mfn2 [132]. A fraction of endogenous Mfn2 is
found in the ER membrane, more precisely at MAMs (see Box 3),
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
10
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
where it is thought to participate in ER–mitochondria tethering by
interacting in trans with Mfn1 and mitochondrial Mfn2. Why the
same proteins are used for both mitochondrial fusion and tethering
to the ER is unclear, as is the fact that Mfn2 can promote ER–
mitochondria tethering without provoking fusion of both organelles
[132]. As tethering is a necessary first step in membrane fusion, a
tethering activity is encoded within mitofusins [133]. Evolution may
thus have reused this property for ER–mitochondria tethering, while
finding a way to prevent unwanted ER–mitochondria fusion.
Whether ER–mitochondria MCSs influence mitochondrial fusion is
not known. In the contrary, fission appears to happen preferentially at
ER–mitochondria MCSs [134]. In a study using both live cell microscopy and electron tomography, the group of Gia Voeltz observed that
events of mitochondrial fission were most frequent at sites of contact
with the ER. In yeast, EM-tomography revealed that MCSs correlated
with sites of slight constriction of the mitochondria. In live mammalian cells, they could see a correlation between intersections of
the ER and the mitochondrial networks and mitochondrial fission
events [134]. Drp1 is recruited to mitochondrial constriction sites by
a mitochondrial-fission-factor (Mff) [135]. Yet a characteristic mitochondrial constriction could also be observed at ER–mitochondria
intersection in the absence of Mff or Drp1, showing that ER–
mitochondria contact is acting upstream of the classical machinery
of mitochondrial fission [134]. What could be the purpose of the coupling between mitochondrial fission and ER–mitochondrial MCSs?
One possibility is that, due to the thinness of the lamellipodia used
in the live imaging experiments [134] (see Box 3), an ER tubule crossing a mitochondrion will mechanically generate a constriction on the
mitochondrion. This mechanical constriction could be the triggering
factor that recruits the mitochondrial fission machinery. More
generally, mechanically induced fission could play a role in resolving
intracellular membrane entanglements. Alternatively, ER contact
could be required to provide factors needed for fission of the mitochondrial membrane. These factors could not only be proteins but
also lipids (see Section 1.2), which may be necessary for the membrane remodeling that underlies fission.
1.3.2.3. Mitochondrial inheritance. During asymmetric cell division,
organelles that cannot be created de novo must be faithfully inherited.
In budding yeast, mitochondrial inheritance is partially dependent on
a Mitochondrial Myo2 Receptor-related protein 1 (Mmr1) [136]. As it
binds the myosin motor Myo2, Mmr1 is thought to promote the
transport of mitochondria along actin cables from the mother to the
bud [136–138]. Yet, MMR1 is also one of a few genes whose messenger RNA is transported into the bud [139]. This suggests that Mmr1 is
required in the bud rather than in the mother, which is inconsistent
with a role in transporting mitochondria from the mother to the
bud. To accommodate this observation, Liza Pon's lab proposes another model: Mmr1 is not required to transport mitochondria to the bud
but instead is required to anchor them to the ER at the bud tip [140].
In such a model, the Myo2-binding activity is only required to localize
Mmr1 to the bud tip, but not to transport mitochondria. Supporting
this model, MMR1 deletion causes a defect in mitochondrial anchoring to the bud tip and an increase in retrograde mitochondrial
movement. This ER–mitochondria contact at the bud tip could be
established by interaction with a fraction of Mmr1 attached to the
ER, substantiated by the presence of a small fraction of Mmr1 in the
MAM fraction [140] (see Box 3).
2. Tethering complexes
The establishment of a functional MCS can be subdivided in a
number of functional steps. The first one is tethering of two membranes. In a second step, effector proteins are recruited to the MCS
to perform Ca 2+ or lipid exchange, or any other function of that particular MCS. However effectors may participate in tethering as much
as tethers could also be effector proteins, which complicates the
search for protein tethers responsible for the establishment of MCSs.
Bona fide tethers should in principle satisfy the following criteria:
1) They should be physically present at MCSs.
2) They should provide a tethering force between adjacent organelles, for example through the physical association of proteins
inserted on opposite membranes.
3) Deletion of a tether should abolish the contacts between two
organelles.
4) Deletion of a tether should affect all physiological processes taking
place at this MCS.
5) Hyperactivation of this tether should increase the number and/or
size of MCSs.
As we will see, very few of the many proposed tethers fulfill all of
these criteria, probably due to the fact that several redundant tethers
exist between a pair of organelles (Table 1), or because MCSs act in
networks where the loss of one MCS can be compensated for by another type of MCS.
2.1. The nuclear—vacuole junction
The yeast NVJ is probably the simplest of all known MCS, since
only two proteins appear to be necessary and sufficient for its establishment. The NVJ protein 1 (NVJ1) on the outer nuclear membrane
interacts with vacuole-related 8 protein (Vac8) on the vacuolar membrane [115,141]. These two proteins alone satisfy all of the criteria
described above for a tether. Deletion of either of these proteins is
sufficient to abolish the NVJ completely, while overexpression increases their extent [115,116]. Finally, PMN, the only well-described
process happening at the NVJ, is crucially dependent on the presence
of both proteins [116,117]. Other proteins are recruited to the NVJ but
do not appear to play a role in tethering per se, such as the ORP Osh1
[93,142], the SMP-containing Nvj2 [105] and the enoyl-CoA reductase
Tsc13 [118,119]. These proteins are thus likely effectors of physiological functions, such as lipid exchange. Interestingly, none of the tethering proteins is conserved, while all of the effectors are. The
mammalian orthologs of at least two of the effectors are likely to be
present at MCSs. ORP1L, a metazoan ortholog of Osh1, harbors both
PH and FFAT domains that direct the protein to ER–late endosome
contact sites, and Testis-EXpressed protein 2 (Tex2), the metazoan
ortholog of Nvj2, has an SMP domain, which has so far only been
found in MCS-proteins [95,105]. Both proteins may thus be found at
MCSs that are analogous to the NVJ, but established and maintained
by other sets of tethers.
2.2. ER–mitochondria tethers
ER–mitochondria contact sites are probably the best understood
MCSs in both lower and higher eukaryotes in terms of the tethering
factors.
2.2.1. The ERMES complex
In budding yeast, a protein complex that tethers the ER and mitochondria was discovered in a forward genetic screen aimed at finding
mutants that can only grow in the presence of exogenous artificial
ER–mitochondria tethering activity [99]. This artificial tether – a GFP
molecule bracketed by mitochondrial and ER targeting sequences
at the N- and C-termini, respectively [51] (see Box 3) – is dispensable
in wildtype cells but becomes indispensable for the respiratory
growth of certain mutants, suggesting that their endogenous tethering capacity has been lost. These mutants are found in genes encoding
members of the ERMES complex, a protein complex made of ER
and mitochondrial proteins, thus satisfying our second criterion. In
accordance to the first criterion, ERMES localizes to few punctate
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
11
Table 1
Summary of the structural and functional players of membrane contact sites.
ER–PM contact sites
Potential tethers
Orai–Stim
Ist2
VAP–ORPs
Tricalbins
Junctophilins
Effectors
Muscle contraction DHPR
RyR
SOCE
Orai
STIM
Lipid exchange
ORPs
Tricalbins
ER–mitochondria contact sites
Potential tethers
ERMES
VDAC–Grp75–IP3R
complex
Mfn2
VAPB–PTPIP51B
Bap31–Fis1
Effectors
Mitochondrial
Ca2+ influx
Lipid exchange
Mitochondrial
dynamics
IP3R
VDAC
MCU
ERMES
Miro
DRP1
Mmr1
ER–vacuole/lysosome contact sites
Potential tethers
Vac8–Nvj1 complex
Effectors
PMN
Lipid exchange
Fatty-acid
synthesis
V-ATPase
Osh1
Nvj2
Tsc13
ER–Golgi contact sites
Potential tethers
VAP–ORPs
VAP–CERT
VAP–PITP
Effectors
Lipid exchange
ORPs
CERT
PITPs
ER–lipid droplets contact sites
Potential tethers
FATP1–DGAT2
ER–late endosome contact sites
Potential tethers
VAP–ORP1L
PTP1B–EGFR
STIM (ER calcium sensor) and Orai (PM calcium channel) form a transient complex upon ER calcium
store depletion. This tether may contribute to tethering the PM to the ER membrane.
ER–membrane protein, can bind PM through a PIP-binding C-terminal domain.
Many ORPs can bind simultaneously to the ER protein VAP, and to PIPs in the PM.
The ER-resident tricalbins 1–3 contain two distinct membrane-binding domains, which can bind the PM.
They localize to ER–PM MCSs.
In muscle cells, SR-resident junctophilins connect the SR to the PM, which they contact through their
lipid-binding MORN-domains.
Section 2.3
Section 2.3
Section 2.3
Section 2.3
Section 2.3.1
DHPR is a voltage-gated PM Ca2+ channel that gates RyR at SR–PM contact sites. RyR is a SR Ca2+ channel.
RyR gating by DHPR amplifies the cytosolic Ca2+ influx during muscle contraction.
STIM is an ER Ca2+ sensor and Orai is a PM Ca2+ channel. Upon Ca2+ depletion in the ER, Stim binds to and
gates Orai at ER–PM MCSs.
ORPs exchange sterols for PI4P. They are often localized at various MCSs.
Tricalbins harbor an SMP domain, suggesting these proteins could act as LTPs.
Section 1.1.1
Complex made of integral ER and OMM proteins. Members of the ERMES complex can be functionally
substituted by an artificial ER–mitochondria tether.
Complex formed by the ER-resident Ca2+ channel IP3R, the cytosolic chaperone Grp75, and the
non-selective channel of the outer membrane VDAC.
An ER-localized subfraction of Mfn2 binds Mfn1/2 in trans on the OMM, thereby tethering the ER to
mitochondria. Mfn2 depletion causes ER fragmentation and Ca2+ overload.
Revealed by a yeast to hybrid screen, this complex consists of the ER protein VAPB and the mitochondrial
protein PTPIP51B. Mutations in VAPB are linked to inheritable forms of multiple sclerosis.
The ER protein Fis1 and the OMM protein Bap31 interact with each other, thereby possibly forming a MCS
between both organelles. The complex seems to function in apoptosis regulation.
Section 2.2.1
IP3Rs are ER-resident ligand-gated channels, which mediate ER Ca2+-release.
OMM general diffusion pore for small hydrophilic molecules.
Mitochondrial Ca2+ uniport responsible for mitochondrial matrix Ca2+ uptake.
Three members of the ERMES complex harbor an SMP domain, suggesting these proteins could act as LTPs.
GTPase involved in mitochondrial movement, found at ER–mitochondria MCSs. Yeast homolog is part of
ERMES complex.
GTPase involved in fission of mitochondrial membranes. Drp1 and mitochondrial fission events are
enriched at ER–mitochondria contact sites.
Yeast myosin binding protein, required for proper transmission of mitochondria to the bud. May be
required to anchor mitochondria to the ER at the bud tip.
Section
Section
Section
Section
Section
Section 1.1.2
Section 1.2.3
Section 1.2.4
Section 2.2.2
Section 2.2.3
Section 2.2.4
Section 2.2.5
1.1.3
1.1.3
1.1.3
1.2.4
1.3.2.1
Section 1.3.2.2
Section 1.3.2.3
Complex composed of vacuolar membrane protein Vac8 and outer nuclear membrane protein Nvj1,
connecting both organelles at nucleus–vacuole junctions (NVJ). Its presence is crucial for establishment
of the NVJ and for downstream processes.
Section 2.1
Involved in formation of the electrochemical gradient across the vacuolar membrane at the NVJ sites
where the PMN takes place.
Enriched at the NVJ. ORPs like Osh1 exchange sterols for PI4P.
Enriched at the NVJ. Harbors an SMP domain, suggesting it could act as LTPs.
A polytopic ER-resident protein enriched at NVJ and required for very-long-chain fatty acids biosynthesis
in yeast.
Section 1.3.1
ORPs, CERTs and PITPs can bind both the ER (via association with VAP) and PIPs in late secretory
organelles, including the Golgi.
Section 2.4
ORPs exchange sterols for PI4P. They are often localized at various MCSs.
CERT extracts ceramide from the ER and transfers it to the Golgi.
PITPs exchange PI for PC between various membranes.
Section 1.2.3
Section 1.2.3
Section 1.2.3
The diacylglycerol acyltransferase 2 (DGAT2) in lipid droplets may interact with the acyl-CoA synthase
(FATP1) in the ER membrane to couple triglycerides synthesis and their deposition in lipid droplets.
Sections 1.2.5
and 2.4
Section 1.2.3
Section 1.2.4
Section 1.3.1
Late-endosome-bound-ORP1L contacts the ER protein VAP in condition of cholesterol depletion in the
Section 2.4
endosome.
PTP1B is an ER-resident phosphatase. It dephosphorylates internalized EGFR. Expression of a phosphatase mutant Section 2.4
increases ER–late-endosome association.
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
12
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
structures at the ER–mitochondria interface where the artificial tether
is also enriched.
Whether ERMES is strictly a tether is a matter of debate (see
Section 1.2.4). On the one hand, it can be substituted by an artificial
tether, suggesting that its function is limited to tethering. On the
other hand, only two of the four essential components of ERMES –
Mdm12 and Mdm34 – can be efficiently replaced by the artificial
tether, showing that the other ERMES components have functions beyond tethering [99]. Yet, deletion of Mdm12 or Mdm34 causes a complete collapse of the ERMES complex, suggesting that these additional
functions are carried out by the remaining ERMES components outside of the complex, therefore outside of MCSs. Alternatively, it is
also possible that the artificial tether helps hold together an otherwise crippled ERMES complex lacking Mdm12 or Mdm34.
2.2.2. The VDAC–GRP75–IP3R complex
In mammalian cells, Ca 2+ transfers mouth-to-mouth from the ER
IP3R to the mitochondrial porin VDAC (voltage dependent anion
channel, see Section 1.1.3.1). Both proteins were actually found to
interact via a cytosolic Hsp70, the glucose-regulated-protein 75
(GRP75) [143]. This indirect interaction of IP3R and VDAC could provide a tethering force between the ER and mitochondria on top of
their role in Ca 2+ homeostasis. In accordance, knockdown of GRP75
decreases ER–mitochondria Ca 2+ exchange.
2.2.3. Mitofusin-2
Mitofusin-2 (Mfn2) is a well-characterized mitochondrial fusion
factor and Mfn2 knockout (KO) cells have fragmented mitochondria.
However, unexpectedly, they also display many ER-related problems.
Their ER is fragmented and has a resting [Ca 2+] nearly twice that of
wildtype. Having an ER phenotype in a mitochondrial mutant may
hint towards a lack of ER–mitochondria tethering. A small fraction
of Mfn2 is found in the MAM fraction, suggesting that it is inserted
in the ER membrane, where it can interact with both Mfn2 and
Mfn1 in the mitochondrial membrane, thereby tethering the two
organelles [132]. Indeed, the ER fragmentation but not the mitochondrial fragmentation of the Mfn2 KO cells can be rescued by expressing
a Mfn2 transgene fused to an ER targeting sequence, suggesting that
this transgene rescues tethering, but not the mitochondrial fusion
function of Mfn2. The nature and the composition of the MAM
fraction (see Box 3) are affected in Mfn2 KO cells, consistent with a
loosening of the ER–mitochondria connection. ER–mitochondria
Ca 2+ transfer is also decreased by ~ 25% [132].
2.2.4. The VAPB–PTPIP51 complex
The ER protein VAP interacts with the FFAT motif found in a
plethora of proteins localized at MCSs between the ER and other membranes, such as the Golgi, the PM and the endo-/lysosome. The VAP
paralog VAPB was further used as bait in a yeast two-hybrid screen,
which led to the identification of a binding partner, the proteintyrosine-phosphatase-interacting protein (PTPIP) 51 [144]. Since
PTPIP51 is an OMM integral protein, PTPIP51–VAPB interaction should
only take place at ER–mitochondria MCSs. In support of this notion, a
fair fraction of VAPB is found in the MAM fraction (see Box 3).
PTPIP51 does not harbor a FFAT motif, thus the interaction between it
and VAPB may happen via other mechanisms. Knocking-down either
VAPB or PTPIP51 causes a ~10% decrease in ER–mitochondrial Ca2+ exchange [144]. Interestingly, a mutation in VAPB (P56S) known to cause
type-8 amyotrophic lateral sclerosis (ALS), shows an increased affinity
for PTPIP51, enrichment in the MAM fraction and mitochondrial
colocalization [144]. Overexpression of this mutant but not of the
wildtype form of VAPB causes a ~1.4 fold increase in ER–mitochondria
Ca2+ exchange. VAPBP56S also decreases the amount of motile mitochondria in the axon by decreasing the association of Miro with microtubules. This decrease can be reversed by expressing a mutant of Miro
that cannot bind Ca2+, suggesting that VAPBP56S acts upon Miro by
increasing [Ca 2+] at the ER–mitochondria MCS [123]. These findings
could bring a new twist to the study of the etiology of ALS.
Although PTPIP51 does not have an FFAT motif, the involvement of
VAPB suggests that VAPs might constitute a hub for several MCSs of
the ER with other cellular organelles.
2.2.5. The Bap31–Fis1 complex
B-cell-receptor-associated protein 31 (Bap31) is an ER protein
that is cleaved by caspases during apoptosis. Fission 1 homologue
(Fis1) is the metazoan ortholog of a yeast OMM protein important
for recruiting dynamin-related protein to mitochondria during mitochondrial fission. While this original role has been assumed by Mff
(see Section 1.3.2.2), metazoan Fis1 acquired a new role in the regulation of apoptosis. Overexpression of Fis1 leads to caspase processing
of Bap31, while knockdown of Fis1 inhibits it. Both proteins interact
physically and constitutively, and they recruit procaspase-8 upon apoptosis induction [145]. This constitutive interaction suggests that
Fis1 and Bap31 could provide a tethering force between the ER and
the mitochondria. However, the function of this complex has not
been studied beyond its role in apoptosis promotion.
2.2.6. Mmr1
As stated in Section 1.3.2.3, yeast Mmr1 may establish an MCS in
the daughter cell to anchor mitochondria to the bud-tip [140]. The
molecular details of this tethering are unclear; in particular, Mmr1's
partner(s) responsible for targeting it to the ER have still to be
identified.
MMR1 shows synthetic genetic interactions with members of the
ERMES complex (2.2.1)[99], suggesting that both contact sites may
have partially overlapping roles.
2.3. ER–plasma-membrane tethers
While many proteins are known to localize either constitutively or
inducibly to ER–PM MCSs, much less is known about the tethering
complexes that establish them. Many of the ER–PM MCS-localized
proteins have a capacity to bind both membranes. Thus many can increase ER–PM association when overexpressed, however, no known
loss-of-function leads to a complete loss of ER–PM tethering.
Lipid exchangers could act as tethers. In yeast, many ORPs are
targeted to ER–PM MCSs (reviewed in [146]). Two of them – Osh2
and Osh3 – have both a FFAT and a PH domain, which could provide
a tethering force by binding to ER VAP proteins (Scs2 and Scs22 in
yeast) and to PM phosphoinositides simultaneously. Another potential
tether comes from the SMP-containing extended-synaptotagmin/
tricalbins proteins (Esyt1-3 in humans, Tcb1-3 in yeast) [105].
Yeast tricalbins accumulate at ER–PM MCS. They are targeted to the
ER via their N-terminal transmembrane domain and interact with
the PM via their C-termini composed of one SMP and repetitions of
C2-domains [147]. They may thus participate in ER–PM tethering.
STIM is engaged in the ER membrane with its transmembrane
sequence. A conformation switch following ER Ca 2 + depletion exposes a C-terminal polybasic stretch that binds to negatively charged
phospholipids in the PM, providing the basis for a transient tether
[23,24]. Indeed, the extent of ER–PM contact is increased upon SOCE
activation [25,40]. Moreover, overexpressing a constitutively active
STIM induces the formation of tethering structures not only between
the ER and the PM, but also between adjacent ER sheets, suggesting
that STIM has an intrinsic tethering activity [40]. Yet, activated STIM
may just extend preexisting structures, instead of creating new
ones. Indeed, successive SOCE activations recruit STIM repeatedly to
the same sites, suggesting that these sites remain in the absence of
STIM [37].
The yeast protein Ist2 is an ER putative ion channel, which, much
like STIM, harbors a C-terminal polybasic domain [148–150]. When
overexpressed in yeast or mammalian cells, Ist2 can increase the
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
13
extent of ER–PM tethering [148,151]. This is particularly spectacular
in yeast where Ist2 overexpression can literally velcro the nuclear envelope against the cell cortex. However, IST2 deletion only has a little
influence on the fact that a large fraction of the ER is in direct contact
with the plasma membrane, suggesting that other tethers must exist.
associated membranes [156] (see Box 3). How DGAT2 translocates from
the ER membrane to the LD outer-layer is completely unknown and
the relative importance of lipid trafficking by this MCS versus lipid trafficking by direct fusion of the ER-membrane with the LD outer-layer
needs to be clarified.
2.3.1. In muscle cells
DHPR and RyR1 physically interact in the membrane junction
complexes. However, this interaction is unlikely to provide the structural basis of these complexes as overexpressing both does not increase the extent of SR–PM MCSs [152]. In an attempt to identify
structural components of junctional membrane complexes (JMCs,
see Section 1.1.1), the group of Hiroshi Takeshima created a library
of monoclonal antibodies raised against muscular membrane proteins, and screened this library for JMC specific epitopes using Electron Microscopy (EM). This approach led to the identification of
junctophilins, a family of proteins with similar architectures [153].
Junctophilins are anchored in the SR via their C-terminal transmembrane domain, while their N-termini consist of repetitions of membrane occupation and recognition nexuses (MORN). The MORN
shows a specific affinity for the PM, therefore providing the basis for
a tether. Mutants of some of the junctophilins show disorganization
of the JMC, while ectopic expression of junctophilins in non-muscle
cells leads to the formation of new ER–PM MCSs. Thus, junctophilins
are likely to contribute to the establishment of SR–PM tethers in
muscles.
3. Conclusions and perspectives
2.4. Other MCSs
Tethering factors involving other cellular structures are much less
characterized. The interaction of VAP proteins with the FFAT domains
of many proteins, such as CERT or ORPs proteins, could contribute to
the tethering of the ER to various organelles [76,77,81]. We already
mentioned the existence of a contact site between the ER and the
late endosome. This contact site may be established by ORP1L via its
FFAT and PH domains in an inducible fashion, depending on the endosome cholesterol content [94,95]. Whether ORP1L is the actual tether is
not clear. Moreover it is not the only candidate for an ER–late endosome tether. Like most plasma-membrane receptor tyrosine kinases,
the epidermal growth factor receptor (EGFR) is endocytosed upon activation and subsequently targeted for degradation in the late endosome/lysosome. On the late endosome, EGFR is dephosphorylated by
the protein tyrosine phosphatase 1B (PTP1B; not to be confused with
PTPIP51, mentioned in Section 2.2.4, nor with PITPs, mentioned in
Section 1.2.1), which is an ER protein [154]. Therefore, the topology
of this dephosphorylation reaction in trans implies the formation of
an ER–late-endosome MCS. When a catalytically inactive form of
PTP1B is expressed, the enzyme–substrate complex cannot be resolved.
Therefore their normally transient interaction becomes durable. As a
result, EM-observable contact sites are created and maintained between the ER and late endosomes, indicating that these two proteins
could be sufficient for the establishment of new MCSs [155]. The interaction of EGFR with wildtype PTP1B may, however, be too transient to
create a real tethering force, thus additional tethers may be involved
(Table 1).
What tethers the ER closely around lipid droplets is unclear. A
recent study proposed that the fatty-acyl Coenzyme A ligase FATP1,
an ER protein that catalyzes the first step of triglyceride biosynthesis,
interacts with the DAG acyltransferase DGAT2 on the surface of LDs,
providing a potential tethering force [114]. This model not only implies that DGAT2 localizes to LDs, but also that it is physically inserted
in the monolayer of the LD. DGAT2 harbors two closely-spaced transmembrane domains with no hydrophilic aminoacid in between them,
thus perfectly fitted to insert into the LD outer-layer. However, DGAT2
is not always colocalized with LDs. It can be seen throughout the ER
in certain conditions and it even cofractionates with mitochondria-
Biochemistry aims to understand biology in terms of chemical
reactions. Yet the cell is far from being a homogenous chemical reactor. Microdomains and subcompartments promote certain reactions
and inhibit others, and organelles play a prime role in this compartmentalization [157]. MCSs are emerging as a new layer of complexity
that allows specific and directional exchange of metabolites and
information between subcompartments. Long confined to phenomenological observations, the biology of MCS has undergone a drastic
speed-up in the recent years with the identification of many factors involved in their structure and/or function. But many questions are still
left unanswered. Are there other molecules besides lipids and Ca 2+
that are specifically exchanged at MCSs? Do the different kinds of
tethers between the same organelles collaborate in establishing one
MCS or does each one establish a specialized MCS linked to a specific
function? How important is non-vesicular lipid transport within the
endomembrane system as compared to vesicular lipid transport?
What LTPs are operating between the ER and the mitochondria?
What is really the MAM fraction? Finally, although the functions
of most contact sites in lipid and Ca 2 + exchange are similar, the factors involved seem unrelated between various MCSs and between
different clades, begging the question of the origin of these structures and of how they have been maintained throughout eukaryotic
evolution.
We can expect that upcoming research will answer these questions and reveal the common organizing principles of this privileged
type of interorganellar communication.
Acknowledgements
We would like to warmly thank Alicia E. Smith for critical comments on the manuscript. This work was supported by grants from
the Swiss National Science Foundation (PP00P3_13365) and from
the ETH (ETH 21 11-2).
References
[1] D.E. Clapham, Calcium signaling, Cell 131 (2007) 1047–1058.
[2] W. Dowhan, Molecular basis for membrane phospholipid diversity: why are
there so many lipids? Annu. Rev. Biochem. 66 (1997) 199–232.
[3] A. Sandow, Excitation–contraction coupling in muscular response, Yale J. Biol.
Med. 25 (1952) 176–201.
[4] F. Brette, C. Orchard, Resurgence of cardiac t-tubule research, Physiology
(Bethesda) 22 (2007) 167–173.
[5] B.A. Block, T. Imagawa, K.P. Campbell, C. Franzini-Armstrong, Structural evidence for direct interaction between the molecular components of the transverse tubule/sarcoplasmic reticulum junction in skeletal muscle, J. Cell Biol.
107 (1988) 2587–2600.
[6] A. Fabiato, Calcium-induced release of calcium from the cardiac sarcoplasmic
reticulum, Am. J. Physiol. 245 (1983) C1–C14.
[7] R.A. Bannister, Bridging the myoplasmic gap: recent developments in skeletal
muscle excitation–contraction coupling, J. Muscle Res. Cell Motil. 28 (2007)
275–283.
[8] K.G. Beam, R.A. Bannister, Looking for answers to EC coupling's persistent questions, J. Gen. Physiol. 136 (2010) 7–12.
[9] R.T. Rebbeck, Y. Karunasekara, E.M. Gallant, P.G. Board, N.A. Beard, M.G.
Casarotto, A.F. Dulhunty, The β(1a) subunit of the skeletal DHPR binds to skeletal RyR1 and activates the channel via its 35-residue C-terminal tail, Biophys. J.
100 (2011) 922–930.
[10] H. Coe, M. Michalak, Calcium binding chaperones of the endoplasmic reticulum,
Gen. Physiol. Biophys. 28 Spec No (2009) F96–F103.
[11] S. Feske, Calcium signalling in lymphocyte activation and disease, Nat. Rev.
Immunol. 7 (2007) 690–702.
[12] J. Liou, M.L. Kim, W. Do Heo, J.T. Jones, J.W. Myers, J.E. Ferrell, T. Meyer, STIM is a
Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx, Curr. Biol.
15 (2005) 1235–1241.
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
14
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
[13] J. Roos, P.J. DiGregorio, A.V. Yeromin, K. Ohlsen, M. Lioudyno, S. Zhang, O. Safrina,
J.A. Kozak, S.L. Wagner, M.D. Cahalan, G. Veliçelebi, K.A. Stauderman, STIM1, an
essential and conserved component of store-operated Ca2+ channel function, J.
Cell Biol. 169 (2005) 435–445.
[14] M. Vig, C. Peinelt, A. Beck, D.L. Koomoa, D. Rabah, M. Koblan-Huberson, S. Kraft,
H. Turner, A. Fleig, R. Penner, J.-P. Kinet, CRACM1 is a plasma membrane protein
essential for store-operated Ca2+ entry, Science 312 (2006) 1220–1223.
[15] S. Feske, Y. Gwack, M. Prakriya, S. Srikanth, S.-H. Puppel, B. Tanasa, P.G. Hogan,
R.S. Lewis, M. Daly, A. Rao, A mutation in Orai1 causes immune deficiency by
abrogating CRAC channel function, Nature 441 (2006) 179–185.
[16] S.L. Zhang, A.V. Yeromin, X.H.-F. Zhang, Y. Yu, O. Safrina, A. Penna, J. Roos, K.a
Stauderman, M.D. Cahalan, Genome-wide RNAi screen of Ca(2+) influx
identifies genes that regulate Ca(2+) release-activated Ca(2+) channel
activity, Proc. Natl. Acad. Sci. U. S. A. 103 (2006) 9357–9362.
[17] A.V. Yeromin, S.L. Zhang, W. Jiang, Y. Yu, O. Safrina, M.D. Cahalan, Molecular
identification of the CRAC channel by altered ion selectivity in a mutant of
Orai, Nature 443 (2006) 226–229.
[18] M. Prakriya, S. Feske, Y. Gwack, S. Srikanth, A. Rao, P.G. Hogan, Orai1 is an essential
pore subunit of the CRAC channel, Nature 443 (2006) 230–233.
[19] J. Liou, M. Fivaz, T. Inoue, T. Meyer, Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion, Proc. Natl. Acad. Sci. U. S. A. 104 (2007)
9301–9306.
[20] P.B. Stathopulos, G.-Y. Li, M.J. Plevin, J.B. Ames, M. Ikura, Stored Ca2+
depletion-induced oligomerization of stromal interaction molecule 1 (STIM1)
via the EF–SAM region: an initiation mechanism for capacitive Ca2+ entry,
J. Biol. Chem. 281 (2006) 35855–35862.
[21] P.B. Stathopulos, L. Zheng, G.-Y. Li, M.J. Plevin, M. Ikura, Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry,
Cell 135 (2008) 110–122.
[22] M. Muik, M. Fahrner, R. Schindl, P. Stathopulos, I. Frischauf, I. Derler, P. Plenk, B.
Lackner, K. Groschner, M. Ikura, C. Romanin, STIM1 couples to ORAI1 via an intramolecular transition into an extended conformation, EMBO J. 30 (2011)
1678–1689.
[23] C.M. Walsh, M. Chvanov, L.P. Haynes, O.H. Petersen, A.V. Tepikin, R.D. Burgoyne,
Role of phosphoinositides in STIM1 dynamics and store-operated calcium entry,
Biochem. J. 425 (2010) 159–168.
[24] M.K. Korzeniowski, M.A. Popovic, Z. Szentpetery, P. Varnai, S.S. Stojilkovic, T.
Balla, Dependence of STIM1/Orai1-mediated calcium entry on plasma membrane phosphoinositides, J. Biol. Chem. 284 (2009) 21027–21035.
[25] M.M. Wu, J. Buchanan, R.M. Luik, R.S. Lewis, Ca2+ store depletion causes STIM1
to accumulate in ER regions closely associated with the plasma membrane,
J. Cell Biol. 174 (2006) 803–813.
[26] S.L. Zhang, Y. Yu, J. Roos, J.A. Kozak, T.J. Deerinck, M.H. Ellisman, K.A. Stauderman,
M.D. Cahalan, STIM1 is a Ca2+ sensor that activates CRAC channels and migrates
from the Ca2+ store to the plasma membrane, Nature 437 (2005) 902–905.
[27] Y. Baba, K. Hayashi, Y. Fujii, A. Mizushima, H. Watarai, M. Wakamori, T. Numaga,
Y. Mori, M. Iino, M. Hikida, T. Kurosaki, Coupling of STIM1 to store-operated
Ca2+ entry through its constitutive and inducible movement in the endoplasmic
reticulum, Proc. Natl. Acad. Sci. U. S. A. 103 (2006) 16704–16709.
[28] R.M. Luik, M.M. Wu, J. Buchanan, R.S. Lewis, The elementary unit of
store-operated Ca2+ entry: local activation of CRAC channels by STIM1 at ER–
plasma membrane junctions, J. Cell Biol. 174 (2006) 815–825.
[29] J.C. Mercer, W.I. Dehaven, J.T. Smyth, B. Wedel, R.R. Boyles, G.S. Bird, J.W. Putney,
Large store-operated calcium selective currents due to co-expression of Orai1 or
Orai2 with the intracellular calcium sensor, Stim1, J. Biol. Chem. 281 (2006)
24979–24990.
[30] P. Xu, J. Lu, Z. Li, X. Yu, L. Chen, T. Xu, Aggregation of STIM1 underneath the
plasma membrane induces clustering of Orai1, Biochem. Biophys. Res. Commun.
350 (2006) 969–976.
[31] T. Kawasaki, I. Lange, S. Feske, A minimal regulatory domain in the C terminus of
STIM1 binds to and activates ORAI1 CRAC channels, Biochem. Biophys. Res.
Commun. 385 (2009) 49–54.
[32] C.Y. Park, P.J. Hoover, F.M. Mullins, P. Bachhawat, E.D. Covington, S. Raunser, T.
Walz, K.C. Garcia, R.E. Dolmetsch, R.S. Lewis, STIM1 clusters and activates
CRAC channels via direct binding of a cytosolic domain to Orai1, Cell 136
(2009) 876–890.
[33] J.P. Yuan, W. Zeng, M.R. Dorwart, Y.-J. Choi, P.F. Worley, S. Muallem, SOAR and
the polybasic STIM1 domains gate and regulate Orai channels, Nat. Cell Biol.
11 (2009) 337–343.
[34] Y. Wang, X. Deng, Y. Zhou, E. Hendron, S. Mancarella, M.F. Ritchie, X.D. Tang, Y.
Baba, T. Kurosaki, Y. Mori, J. Soboloff, D.L. Gill, STIM protein coupling in the activation of Orai channels, Proc. Natl. Acad. Sci. U. S. A. 106 (2009) 7391–7396.
[35] M. Muik, M. Fahrner, I. Derler, R. Schindl, J. Bergsmann, I. Frischauf, K. Groschner,
C. Romanin, A cytosolic homomerization and a modulatory domain within
STIM1 C terminus determine coupling to ORAI1 channels, J. Biol. Chem. 284
(2009) 8421–8426.
[36] Y. Zhou, P. Meraner, H.T. Kwon, D. Machnes, M. Oh-hora, J. Zimmer, Y. Huang, A.
Stura, A. Rao, P.G. Hogan, STIM1 gates the store-operated calcium channel ORAI1
in vitro, Nat. Struct. Mol. Biol. 17 (2010) 112–116.
[37] J.T. Smyth, W.I. Dehaven, G.S. Bird, J.W. Putney, Ca2+-store-dependent and
-independent reversal of Stim1 localization and function, J. Cell Sci. 121
(2008) 762–772.
[38] M. Chvanov, C.M. Walsh, L.P. Haynes, S.G. Voronina, G. Lur, O.V. Gerasimenko, R.
Barraclough, P.S. Rudland, O.H. Petersen, R.D. Burgoyne, A.V. Tepikin, ATP depletion induces translocation of STIM1 to puncta and formation of STIM1-ORAI1
[39]
[40]
[41]
[42]
[43]
[44]
[45]
[46]
[47]
[48]
[49]
[50]
[51]
[52]
[53]
[54]
[55]
[56]
[57]
[58]
[59]
[60]
[61]
[62]
[63]
[64]
[65]
[66]
clusters: translocation and re-translocation of STIM1 does not require ATP,
Pflügers Archiv/: European, J. Physiol. 457 (2008) 505–517.
G. Lur, L.P. Haynes, I.A. Prior, O.V. Gerasimenko, S. Feske, O.H. Petersen, R.D.
Burgoyne, A.V. Tepikin, Ribosome-free terminals of rough ER allow formation
of STIM1 puncta and segregation of STIM1 from IP(3) receptors, Curr. Biol. 19
(2009) 1648–1653.
L. Orci, M. Ravazzola, M. Le Coadic, W.-W. Shen, N. Demaurex, P. Cosson, From the
cover: STIM1-induced precortical and cortical subdomains of the endoplasmic
reticulum, Proc. Natl. Acad. Sci. U. S. A. 106 (2009) 19358–19362.
H. Jousset, M. Frieden, N. Demaurex, STIM1 knockdown reveals that
store-operated Ca2+ channels located close to sarco/endoplasmic Ca2+ ATPases
(SERCA) pumps silently refill the endoplasmic reticulum, J. Biol. Chem. 282
(2007) 11456–11464.
I.M. Manjarrés, M.T. Alonso, J. García-Sancho, Calcium entry-calcium refilling
(CECR) coupling between store-operated Ca(2+) entry and sarco/endoplasmic
reticulum Ca(2+)-ATPase, Cell Calcium 49 (2011) 153–161.
A. Sampieri, A. Zepeda, A. Asanov, L. Vaca, Visualizing the store-operated
channel complex assembly in real time: identification of SERCA2 as a new
member, Cell Calcium 45 (2009) 439–446.
E. Carafoli, The interplay of mitochondria with calcium: an historical appraisal,
Cell Calcium 52 (2012) 1–8.
J.M. Baughman, F. Perocchi, H.S. Girgis, M. Plovanich, C.A. Belcher-Timme, Y.
Sancak, X.R. Bao, L. Strittmatter, O. Goldberger, R.L. Bogorad, V. Koteliansky,
V.K. Mootha, Integrative genomics identifies MCU as an essential component
of the mitochondrial calcium uniporter, Nature 476 (2011) 341–345.
D. De Stefani, A. Raffaello, E. Teardo, I. Szabò, R. Rizzuto, A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter, Nature 476
(2011) 336–340.
R. Rizzuto, M. Brini, M. Murgia, T. Pozzan, Microdomains with high Ca2+ close to
IP3-sensitive channels that are sensed by neighboring mitochondria, Science
262 (1993) 744–747.
R. Rizzuto, P. Pinton, W. Carrington, F.S. Fay, K.E. Fogarty, L.M. Lifshitz, R.A. Tuft,
T. Pozzan, Close contacts with the endoplasmic reticulum as determinants of
mitochondrial Ca2+ responses, Science 280 (1998) 1763–1766.
G. Csordás, P. Várnai, T. Golenár, S. Roy, G. Purkins, T.G. Schneider, T. Balla, G.
Hajnóczky, Imaging interorganelle contacts and local calcium dynamics at the
ER–mitochondrial interface, Mol. Cell 39 (2010) 121–132.
M. Giacomello, I. Drago, M. Bortolozzi, M. Scorzeto, A. Gianelle, P. Pizzo, T.
Pozzan, Ca2+ hot spots on the mitochondrial surface are generated by Ca2+ mobilization from stores, but not by activation of store-operated Ca2+ channels,
Mol. Cell 38 (2010) 280–290.
G. Csordás, C. Renken, P. Várnai, L. Walter, D. Weaver, K.F. Buttle, T. Balla, C.A.
Mannella, G. Hajnóczky, Structural and functional features and significance of
the physical linkage between ER and mitochondria, J. Cell Biol. 174 (2006)
915–921.
R.M. Denton, Regulation of mitochondrial dehydrogenases by calcium ions,
Biochim. Biophys. Acta 1787 (2009) 1309–1316.
A.I. Tarasov, E.J. Griffiths, G.A. Rutter, Regulation of ATP production by mitochondrial Ca(2+), Cell Calcium 52 (2012) 28–35.
C. Cárdenas, R.A. Miller, I. Smith, T. Bui, J. Molgó, M. Müller, H. Vais, K.-H. Cheung,
J. Yang, I. Parker, C.B. Thompson, M.J. Birnbaum, K.R. Hallows, J.K. Foskett, Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer
to mitochondria, Cell 142 (2010) 270–283.
G. Hajnóczky, R. Hager, A.P. Thomas, Mitochondria suppress local feedback
activation of inositol 1,4,5-trisphosphate receptors by Ca2+, J. Biol. Chem. 274
(1999) 14157–14162.
E. Boitier, R. Rea, M.R. Duchen, Mitochondria exert a negative feedback on the
propagation of intracellular Ca2+ waves in rat cortical astrocytes, J. Cell Biol.
145 (1999) 795–808.
R.A. Haworth, D.R. Hunter, The Ca2+-induced membrane transition in mitochondria II Nature of the Ca2+ trigger site, Arch. Biochem. Biophys. 195 (1979)
460–467.
P. Bernardi, A. Rasola, Calcium and cell death: the mitochondrial connection,
Subcell. Biochem. 45 (2007) 481–506.
P. Pinton, D. Ferrari, P. Magalhães, K. Schulze-Osthoff, F. Di Virgilio, T. Pozzan, R.
Rizzuto, Reduced loading of intracellular Ca(2+) stores and downregulation of
capacitative Ca(2 +) influx in Bcl-2-overexpressing cells, J. Cell Biol. 148 (2000)
857–862.
L. Scorrano, S.A. Oakes, J.T. Opferman, E.H. Cheng, M.D. Sorcinelli, T. Pozzan, S.J.
Korsmeyer, BAX and BAK regulation of endoplasmic reticulum Ca2+: a control
point for apoptosis, Science 300 (2003) 135–139.
M. Chami, B. Oulès, G. Szabadkai, R. Tacine, R. Rizzuto, P. Paterlini-Bréchot, Role
of SERCA1 truncated isoform in the proapoptotic calcium transfer from ER to
mitochondria during ER stress, Mol. Cell 32 (2008) 641–651.
T. Jayaraman, A. Marks, T cells deficient in inositol 1,4,5-trisphosphate receptor
are resistant to apoptosis, Mol. Cell. Biol. 17 (1997) 3005–3012.
H. Sugawara, M. Kurosaki, M. Takata, T. Kurosaki, Genetic evidence for involvement of type 1, type 2 and type 3 inositol 1,4,5-trisphosphate receptors in signal transduction through the B-cell antigen receptor, EMBO J. 16 (1997)
3078–3088.
C.R.H. Raetz, in: J.N.H., G.B.A.B.T.-N.C. (Eds.), Chapter 11 Genetic Control of
Phospholipid Bilayer Assembly, BiochemistryElsevier, 1982, pp. 435–477.
R.G. Sleight, Intracellular lipid transport in eukaryotes, Annu. Rev. Physiol. 49
(1987) 193–208.
G. van Meer, D.R. Voelker, G.W. Feigenson, Membrane lipids: where they are and
how they behave, Nat. Rev. Mol. Cell Biol. 9 (2008) 112–124.
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
[67] G. van Meer, A.I.P.M. de Kroon, Lipid map of the mammalian cell, J. Cell Sci. 124
(2011) 5–8.
[68] J.E. Vance, E.J. Aasman, R. Szarka, Brefeldin A does not inhibit the movement of
phosphatidylethanolamine from its sites for synthesis to the cell surface,
J. Biol. Chem. 266 (1991) 8241–8247.
[69] J.W. Kok, T. Babia, K. Klappe, G. Egea, D. Hoekstra, Ceramide transport from endoplasmic reticulum to Golgi apparatus is not vesicle-mediated, Biochem. J. 333
(Pt 3) (1998) 779–786.
[70] L. Urbani, R.D. Simoni, Cholesterol and vesicular stomatitis virus G protein take
separate routes from the endoplasmic reticulum to the plasma membrane,
J. Biol. Chem. 265 (1990) 1919–1923.
[71] C. Osman, D.R. Voelker, T. Langer, Making heads or tails of phospholipids in
mitochondria, J. Cell Biol. 192 (2011) 7–16.
[72] J.E. Vance, Phospholipid synthesis in a membrane fraction associated with
mitochondria, J. Biol. Chem. 265 (1990) 7248–7256.
[73] S. Lev, Non-vesicular lipid transport by lipid-transfer proteins and beyond, Nat.
Rev. Mol. Cell Biol. 11 (2010) 739–750.
[74] A. Toulmay, W.A. Prinz, Lipid transfer and signaling at organelle contact sites:
the tip of the iceberg, Curr. Opin. Cell Biol. 23 (2011) 458–463.
[75] K. Hanada, Discovery of the molecular machinery CERT for endoplasmic
reticulum-to-Golgi trafficking of ceramide, Mol. Cell. Biochem. 286 (2006) 23–31.
[76] C.J.R. Loewen, A. Roy, T.P. Levine, A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP, EMBO J. 22 (2003)
2025–2035.
[77] D. Peretti, N. Dahan, E. Shimoni, K. Hirschberg, S. Lev, Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins
and is essential for Golgi-mediated transport, Mol. Biol. Cell 19 (2008) 3871–3884.
[78] R.J. Haslam, H.B. Koide, B.A. Hemmings, Pleckstrin domain homology, Nature
363 (1993) 309–310.
[79] M.A. Lemmon, K.M. Ferguson, Molecular determinants in pleckstrin homology
domains that allow specific recognition of phosphoinositides, Biochem. Soc.
Trans. 29 (2001) 377–384.
[80] B.J. Mayer, R. Ren, K.L. Clark, D. Baltimore, A putative modular domain present in
diverse signalling molecules, Cell 73 (1993) 629–630.
[81] M.J. Rebecchi, S. Scarlata, Pleckstrin homology domains: a common fold with
diverse functions, Annu. Rev. Biophys. Biomol. Struct. 27 (1998) 503–528.
[82] G. Drin, J.-F. Casella, R. Gautier, T. Boehmer, T.U. Schwartz, B. Antonny, A general
amphipathic alpha-helical motif for sensing membrane curvature, Nat. Struct.
Mol. Biol. 14 (2007) 138–146.
[83] M.D. Yoder, L.M. Thomas, J.M. Tremblay, R.L. Oliver, L.R. Yarbrough, G.M.
Helmkamp, Structure of a multifunctional protein mammalian phosphatidylinositol transfer protein complexed with phosphatidylcholine, J. Biol.
Chem. 276 (2001) 9246–9252.
[84] S.J. Tilley, A. Skippen, J. Murray-Rust, P.M. Swigart, A. Stewart, C.P. Morgan, S.
Cockcroft, N.Q. McDonald, Structure-function analysis of human [corrected]
phosphatidylinositol transfer protein alpha bound to phosphatidylinositol,
Structure 12 (2004) 317–326.
[85] N. Kudo, K. Kumagai, N. Tomishige, T. Yamaji, S. Wakatsuki, M. Nishijima, K.
Hanada, R. Kato, Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide, Proc. Natl. Acad. Sci. U. S. A. 105
(2008) 488–493.
[86] Y.J. Im, S. Raychaudhuri, W.A. Prinz, J.H. Hurley, Structural mechanism for sterol
sensing and transport by OSBP-related proteins, Nature 437 (2005) 154–158.
[87] M. de Saint-Jean, V. Delfosse, D. Douguet, G. Chicanne, B. Payrastre, W. Bourguet,
B. Antonny, G. Drin, Osh4p exchanges sterols for phosphatidylinositol
4-phosphate between lipid bilayers, J. Cell Biol. 195 (2011) 965–978.
[88] C.T. Beh, C.R. McMaster, K.G. Kozminski, A.K. Menon, A detour for yeast oxysterol
binding proteins, J. Biol. Chem. 287 (2012) 11481–11488.
[89] C.J. Stefan, A.G. Manford, D. Baird, J. Yamada-Hanff, Y. Mao, S.D. Emr, Osh proteins regulate phosphoinositide metabolism at ER–plasma membrane contact
sites, Cell 144 (2011) 389–401.
[90] V.A. Bankaitis, J.R. Aitken, A.E. Cleves, W. Dowhan, An essential role for a
phospholipid transfer protein in yeast Golgi function, Nature 347 (1990)
561–562.
[91] K. Huitema, J. van den Dikkenberg, J.F.H.M. Brouwers, J.C.M. Holthuis, Identification of a family of animal sphingomyelin synthases, EMBO J. 23 (2004) 33–44.
[92] C.L. Baron, V. Malhotra, Role of diacylglycerol in PKD recruitment to the TGN and
protein transport to the plasma membrane, Science 295 (2002) 325–328.
[93] T.P. Levine, S. Munro, Dual targeting of Osh1p, a yeast homologue of
oxysterol-binding protein, to both the Golgi and the nucleus–vacuole junction,
Mol. Biol. Cell 12 (2001) 1633–1644.
[94] M. Johansson, N. Rocha, W. Zwart, I. Jordens, L. Janssen, C. Kuijl, V.M. Olkkonen, J.
Neefjes, Activation of endosomal dynein motors by stepwise assembly of
Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin, J. Cell Biol. 176
(2007) 459–471.
[95] M. Johansson, M. Lehto, K. Tanhuanpa, T.L. Cover, V.M. Olkkonen, The
oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters
functional properties of late endocytic compartments, Biol. Cell 16 (2005)
5480–5492.
[96] N. Rocha, C. Kuijl, R. van der Kant, L. Janssen, D. Houben, H. Janssen, W. Zwart, J.
Neefjes, Cholesterol sensor ORP1L contacts the ER protein VAP to control
Rab7-RILP-p150 Glued and late endosome positioning, J. Cell Biol. 185 (2009)
1209–1225.
[97] P. Wang, W. Duan, A.L. Munn, H. Yang, Molecular characterization of Osh6p, an
oxysterol binding protein homolog in the yeast Saccharomyces cerevisiae, FEBS J.
272 (2005) 4703–4715.
15
[98] T.A. Schulz, M.-G. Choi, S. Raychaudhuri, J.A. Mears, R. Ghirlando, J.E. Hinshaw,
W.A. Prinz, Lipid-regulated sterol transfer between closely apposed membranes
by oxysterol-binding protein homologues, J. Cell Biol. 187 (2009) 889–903.
[99] B. Kornmann, E. Currie, S.R. Collins, M. Schuldiner, J. Nunnari, J.S. Weissman, P.
Walter, An ER–mitochondria tethering complex revealed by a synthetic biology
screen, Science 325 (2009) 477–481.
[100] B. Kornmann, C. Osman, P. Walter, The conserved GTPase Gem1 regulates endoplasmic reticulum–mitochondria connections, Proc. Natl. Acad. Sci. U. S. A. 108
(2011) 14151–14156.
[101] D.A. Stroud, S. Oeljeklaus, S. Wiese, M. Bohnert, U. Lewandrowski, A. Sickmann,
B. Guiard, M. van der Laan, B. Warscheid, N. Wiedemann, Composition and topology of the endoplasmic reticulum–mitochondria encounter structure, J.
Mol. Biol. 413 (2011) 743–750.
[102] B. Kornmann, P. Walter, ERMES-mediated ER–mitochondria contacts: molecular
hubs for the regulation of mitochondrial biology, J. Cell Sci. 123 (2010)
1389–1393.
[103] I. Lee, W. Hong, Diverse membrane-associated proteins contain a novel SMP
domain, FASEB J. 20 (2006) 202–206.
[104] K.O. Kopec, V. Alva, A.N. Lupas, Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange
between ER and mitochondria, Bioinformatics 26 (2010) 1927–1931.
[105] A. Toulmay, W.A. Prinz, A conserved membrane-binding domain targets proteins to organelle contact sites, J. Cell Sci. 125 (2012) 49–58.
[106] C. Voss, S. Lahiri, B.P. Young, C.J. Loewen, W.A. Prinz, ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae, J. Cell Sci.
125 (2012) 4791–4799.
[107] T.T. Nguyen, A. Lewandowska, J.-Y. Choi, D.F. Markgraf, M. Junker, M. Bilgin, C.S.
Ejsing, D.R. Voelker, T.A. Rapoport, J.M. Shaw, Gem1 and ERMES do not directly
affect phosphatidylserine transport from ER to mitochondria or mitochondrial
inheritance, Traffic 13 (2012) 880–890.
[108] T.C. Walther, R.V. Farese, Lipid droplets and cellular lipid metabolism, Annu. Rev.
Biochem. 81 (2012) 687–714.
[109] S.L. Sturley, M.M. Hussain, Lipid droplet formation on opposing sides of the
endoplasmic reticulum, J. Lipid Res. 53 (2012) 1800–1810.
[110] N. Jacquier, V. Choudhary, M. Mari, A. Toulmay, F. Reggiori, R. Schneiter, Lipid
droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae, J. Cell Sci. 124 (2011) 2424–2437.
[111] E.J. Blanchette-Mackie, N.K. Dwyer, T. Barber, R.A. Coxey, T. Takeda, C.M.
Rondinone, J.L. Theodorakis, A.S. Greenberg, C. Londos, Perilipin is located on
the surface layer of intracellular lipid droplets in adipocytes, J. Lipid Res. 36
(1995) 1211–1226.
[112] H. Wolinski, D. Kolb, S. Hermann, R.I. Koning, S.D. Kohlwein, A role for seipin in
lipid droplet dynamics and inheritance in yeast, J. Cell Sci. 124 (2011)
3894–3904.
[113] H. Robenek, O. Hofnagel, I. Buers, M.J. Robenek, D. Troyer, N.J. Severs,
Adipophilin-enriched domains in the ER membrane are sites of lipid droplet
biogenesis, J. Cell Sci. 119 (2006) 4215–4224.
[114] N. Xu, S.O. Zhang, R.A. Cole, S.A. McKinney, F. Guo, J.T. Haas, S. Bobba, R.V. Farese,
H.Y. Mak, The FATP1–DGAT2 complex facilitates lipid droplet expansion at the
ER–lipid droplet interface, J. Cell Biol. 198 (2012) 895–911.
[115] X. Pan, P. Roberts, Y. Chen, E. Kvam, N. Shulga, K. Huang, S. Lemmon, D.S.
Goldfarb, Nucleus–vacuole junctions in Saccharomyces cerevisiae are formed
through the direct interaction of Vac8p with Nvj1p, Mol. Biol. Cell 11 (2000)
2445–2457.
[116] P. Roberts, S. Moshitch-Moshkovitz, E. Kvam, E. O'Toole, M. Winey, D.S. Goldfarb,
Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae, Mol. Biol. Cell
14 (2003) 129–141.
[117] R. Dawaliby, A. Mayer, Microautophagy of the nucleus coincides with a vacuolar
diffusion barrier at nuclear–vacuolar junctions, Mol. Biol. Cell 21 (2010)
4173–4183.
[118] S.D. Kohlwein, S. Eder, C.S. Oh, C.E. Martin, K. Gable, D. Bacikova, T. Dunn, Tsc13p
is required for fatty acid elongation and localizes to a novel structure at the
nuclear-vacuolar interface in Saccharomyces cerevisiae, Mol. Cell. Biol. 21
(2001) 109–125.
[119] E. Kvam, K. Gable, T.M. Dunn, D.S. Goldfarb, Targeting of Tsc13p to nucleus–
vacuole junctions: a role for very-long-chain fatty acids in the biogenesis of
microautophagic vesicles, Mol. Biol. Cell 16 (2005) 3987–3998.
[120] J.R. Friedman, B.M. Webster, D.N. Mastronarde, K.J. Verhey, G.K. Voeltz, ER sliding dynamics and ER–mitochondrial contacts occur on acetylated microtubules,
J. Cell Biol. 190 (2010) 363–375.
[121] X. Guo, G.T. Macleod, A. Wellington, F. Hu, S. Panchumarthi, M. Schoenfield, L.
Marin, M.P. Charlton, H.L. Atwood, K.E. Zinsmaier, The GTPase dMiro is required
for axonal transport of mitochondria to Drosophila synapses, Neuron 47 (2005)
379–393.
[122] X. Wang, T.L. Schwarz, The mechanism of Ca2+-dependent regulation of
kinesin-mediated mitochondrial motility, Cell 136 (2009) 163–174.
[123] G.M. Mórotz, K.J. De Vos, A. Vagnoni, S. Ackerley, C.E. Shaw, C.C.J. Miller,
Amyotrophic lateral sclerosis-associated mutant VAPBP56S perturbs calcium
homeostasis to disrupt axonal transport of mitochondria, Hum. Mol. Genet. 21
(2012) 1979–1988.
[124] G.J. Hermann, J.W. Thatcher, J.P. Mills, K.G. Hales, M.T. Fuller, J. Nunnari, J.M.
Shaw, Mitochondrial fusion in yeast requires the transmembrane GTPase
Fzo1p, J. Cell Biol. 143 (1998) 359–373.
[125] W. Bleazard, J.M. McCaffery, E.J. King, S. Bale, A. Mozdy, Q. Tieu, J. Nunnari, J.M.
Shaw, The dynamin-related GTPase Dnm1 regulates mitochondrial fission in
yeast, Nat. Cell Biol. 1 (1999) 298–304.
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028
16
S.C.J. Helle et al. / Biochimica et Biophysica Acta xxx (2013) xxx–xxx
[126] E. Smirnova, L. Griparic, D.L. Shurland, A.M. van der Bliek, Dynamin-related
protein Drp1 is required for mitochondrial division in mammalian cells, Mol.
Biol. Cell 12 (2001) 2245–2256.
[127] N. Ishihara, Y. Eura, K. Mihara, Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity, J. Cell Sci. 117 (2004) 6535–6546.
[128] E. Ingerman, E.M. Perkins, M. Marino, J.A. Mears, J.M. McCaffery, J.E. Hinshaw, J.
Nunnari, Dnm1 forms spirals that are structurally tailored to fit mitochondria,
J. Cell Biol. 170 (2005) 1021–1027.
[129] D. Otsuga, B.R. Keegan, E. Brisch, J.W. Thatcher, G.J. Hermann, W. Bleazard, J.M.
Shaw, The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast, J. Cell Biol. 143 (1998) 333–349.
[130] A.M. Labrousse, M.D. Zappaterra, D.A. Rube, A.M. van der Bliek, C elegans
dynamin-related protein DRP-1 controls severing of the mitochondrial outer
membrane, Mol. Cell 4 (1999) 815–826.
[131] H. Chen, S.A. Detmer, A.J. Ewald, E.E. Griffin, S.E. Fraser, D.C. Chan, Mitofusins
Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential
for embryonic development, J. Cell Biol. 160 (2003) 189–200.
[132] O.M. de Brito, L. Scorrano, Mitofusin 2 tethers endoplasmic reticulum to mitochondria, Nature 456 (2008) 605–610.
[133] T. Koshiba, S.A. Detmer, J.T. Kaiser, H. Chen, J.M. McCaffery, D.C. Chan, Structural basis
of mitochondrial tethering by mitofusin complexes, Science 305 (2004) 858–862.
[134] J.R. Friedman, L.L. Lackner, M. West, J.R. DiBenedetto, J. Nunnari, G.K. Voeltz, ER
tubules mark sites of mitochondrial division, Science 334 (2011) 358–362.
[135] H. Otera, C. Wang, M.M. Cleland, K. Setoguchi, S. Yokota, R.J. Youle, K. Mihara,
Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells, J. Cell Biol. 191 (2010) 1141–1158.
[136] T. Itoh, A. Toh-E, Y. Matsui, Mmr1p is a mitochondrial factor for Myo2p-dependent
inheritance of mitochondria in the budding yeast, EMBO J. 23 (2004) 2520–2530.
[137] R.L. Frederick, K. Okamoto, J.M. Shaw, Multiple pathways influence mitochondrial inheritance in budding yeast, Genetics 178 (2008) 825–837.
[138] V.R. Simon, T.C. Swayne, L.A. Pon, Actin-dependent mitochondrial motility in
mitotic yeast and cell-free systems: identification of a motor activity on the
mitochondrial surface, J. Cell Biol. 130 (1995) 345–354.
[139] K.A. Shepard, A.P. Gerber, A. Jambhekar, P.A. Takizawa, P.O. Brown, D. Herschlag,
J.L. DeRisi, R.D. Vale, Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis, Proc. Natl.
Acad. Sci. U. S. A. 100 (2003) 11429–11434.
[140] T.C. Swayne, C. Zhou, I.R. Boldogh, J.K. Charalel, J.R. McFaline-Figueroa, S. Thoms,
C. Yang, G. Leung, J. McInnes, R. Erdmann, L.a Pon, I. Boldogh, L.A. Pon, Role for
cER and Mmr1p in anchorage of mitochondria at sites of polarized surface
growth in budding yeast, Curr. Biol. 21 (2011) 1994–1999.
[141] J.I. Millen, J. Pierson, E. Kvam, L.J. Olsen, D.S. Goldfarb, The luminal N-terminus of
yeast Nvj1 is an inner nuclear membrane anchor, Traffic 9 (2008) 1653–1664.
[142] E. Kvam, D.S. Goldfarb, Nvj1p is the outer-nuclear-membrane receptor for
oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae, J. Cell
Sci. 117 (2004) 4959–4968.
[143] G. Szabadkai, K. Bianchi, P. Várnai, D. De Stefani, M.R. Wieckowski, D. Cavagna,
A.I. Nagy, T. Balla, R. Rizzuto, Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels, J. Cell Biol. 175 (2006) 901–911.
[144] K.J. De Vos, G.M. Mórotz, R. Stoica, E.L. Tudor, K.-F. Lau, S. Ackerley, A. Warley,
C.E. Shaw, C.C.J. Miller, VAPB interacts with the mitochondrial protein PTPIP51
to regulate calcium homeostasis, Hum. Mol. Genet. 21 (2012) 1299–1311.
[145] R. Iwasawa, A.-L. Mahul-Mellier, C. Datler, E. Pazarentzos, S. Grimm, Fis1 and
Bap31 bridge the mitochondria–ER interface to establish a platform for apoptosis induction, EMBO J. 30 (2011) 556–568.
[146] S. Raychaudhuri, W.A. Prinz, The diverse functions of oxysterol-binding proteins, Annu. Rev. Cell Dev. Biol. 26 (2010) 157–177.
[147] C.E. Creutz, S.L. Snyder, T.A. Schulz, Characterization of the yeast tricalbins:
membrane-bound multi-C2-domain proteins that form complexes involved in
membrane trafficking, Cell. Mol. Life Sci. 61 (2004) 1208–1220.
[148] W. Wolf, A. Kilic, B. Schrul, H. Lorenz, B. Schwappach, M. Seedorf, Yeast Ist2 recruits the endoplasmic reticulum to the plasma membrane and creates a
ribosome-free membrane microcompartment, PLoS One 7 (2012) e39703.
[149] K. Maass, M.A. Fischer, M. Seiler, K. Temmerman, W. Nickel, M. Seedorf, A signal comprising a basic cluster and an amphipathic alpha–helix interacts with lipids and is required for the transport of Ist2 to the yeast cortical ER, J. Cell Sci. 122 (2009) 625–635.
[150] E. Ercan, F. Momburg, U. Engel, K. Temmerman, W. Nickel, M. Seedorf, A conserved, lipid-mediated sorting mechanism of yeast Ist2 and mammalian STIM
proteins to the peripheral ER, Traffic 10 (2009) 1802–1818.
[151] G. Lavieu, L. Orci, L. Shi, M. Geiling, M. Ravazzola, F. Wieland, P. Cosson, J.E.
Rothman, Induction of cortical endoplasmic reticulum by dimerization of a
coatomer-binding peptide anchored to endoplasmic reticulum membranes,
Proc. Natl. Acad. Sci. U. S. A. 107 (2010) 6876–6881.
[152] H. Takekura, H. Takeshima, S. Nishimura, M. Takahashi, T. Tanabe, V. Flockerzi, F.
Hofmann, C. Franzini-Armstrong, Co-expression in CHO cells of two muscle proteins involved in excitation–contraction coupling, J. Muscle Res. Cell Motil. 16
(1995) 465–480.
[153] H. Takeshima, S. Komazaki, M. Nishi, M. Iino, K. Kangawa, Junctophilins: a novel
family of junctional membrane complex proteins, Mol. Cell 6 (2000) 11–22.
[154] F.G. Haj, P.J. Verveer, A. Squire, B.G. Neel, P.I.H. Bastiaens, Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum,
Science 295 (2002) 1708–1711.
[155] E.R. Eden, I.J. White, A. Tsapara, C.E. Futter, Membrane contacts between
endosomes and ER provide sites for PTP1B-epidermal growth factor receptor
interaction, Nat. Cell Biol. 12 (2010) 267–272.
[156] S.J. Stone, M.C. Levin, P. Zhou, J. Han, T.C. Walther, R.V. Farese, The endoplasmic
reticulum enzyme DGAT2 is found in mitochondria-associated membranes and
has a mitochondrial targeting signal that promotes its association with mitochondria, J. Biol. Chem. 284 (2009) 5352–5361.
[157] J.C.M. Holthuis, C. Ungermann, Cellular microcompartments constitute general
sub-organellar functional units in cells, Biol. Chem. 394 (2012) 151–161.
[158] A.P. Gasch, P.T. Spellman, C.M. Kao, O. Carmel-Harel, M.B. Eisen, G. Storz, D.
Botstein, P.O. Brown, Genomic expression programs in the response of yeast
cells to environmental changes, Mol. Biol. Cell 11 (2000) 4241–4257.
[159] K. Simons, E. Ikonen, Functional rafts in cell membranes, Nature 387 (1997)
569–572.
[160] D. Lingwood, K. Simons, Detergent resistance as a tool in membrane research,
Nat. Protoc. 2 (2007) 2159–2165.
[161] S. Munro, Lipid rafts: elusive or illusive? Cell 115 (2003) 377–388.
[162] T. Hayashi, M. Fujimoto, Detergent-resistant microdomains determine the
localization of sigma-1 receptors to the endoplasmic reticulum–mitochondria
junction, Mol. Pharmacol. 77 (2010) 517–528.
[163] E. Area-Gomez, M. Del Carmen Lara Castillo, M.D. Tambini, C. Guardia-Laguarta,
A.J.C. de Groof, M. Madra, J. Ikenouchi, M. Umeda, T.D. Bird, S.L. Sturley, E.A.
Schon, Upregulated function of mitochondria-associated ER membranes in
Alzheimer disease, EMBO J. 31 (2012) 4106–4123.
[164] M. Fujimoto, T. Hayashi, T.-P. Su, The role of cholesterol in the association of
endoplasmic reticulum membranes with mitochondria, Biochem. Biophys. Res.
Commun. 417 (2012) 635–639.
[165] T. Hayashi, T.-P. Su, Sigma-1 receptors at galactosylceramide-enriched lipid
microdomains regulate oligodendrocyte differentiation, Proc. Natl. Acad. Sci. U.
S. A. 101 (2004) 14949–14954.
[166] B. Pani, H.L. Ong, X. Liu, K. Rauser, I.S. Ambudkar, B.B. Singh, Lipid rafts determine clustering of STIM1 in endoplasmic reticulum–plasma membrane junctions and regulation of store-operated Ca2+ entry (SOCE), J. Biol. Chem. 283
(2008) 17333–17340.
[167] A.E. Rusiñol, Z. Cui, M.H. Chen, J.E. Vance, A unique mitochondria-associated
membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins, J. Biol. Chem.
269 (1994) 27494–27502.
[168] H. Pichler, B. Gaigg, C. Hrastnik, G. Achleitner, S.D. Kohlwein, G. Zellnig, A.
Perktold, G. Daum, A subfraction of the yeast endoplasmic reticulum associates
with the plasma membrane and has a high capacity to synthesize lipids, Eur. J.
Biochem. 268 (2001) 2351–2361.
[169] K. Kozieł, M. Lebiedzinska, G. Szabadkai, M. Onopiuk, W. Brutkowski, K.
Wierzbicka, G. Wilczyński, P. Pinton, J. Duszyński, K. Zabłocki, M.R.
Wieckowski, Plasma membrane associated membranes (PAM) from Jurkat
cells contain STIM1 protein: is PAM involved in the capacitative calcium
entry? Int. J. Biochem. Cell Biol. 41 (2009) 2440–2449.
[170] M.X. Andersson, M. Goksör, A.S. Sandelius, Optical manipulation reveals strong
attracting forces at membrane contact sites between endoplasmic reticulum
and chloroplasts, J. Biol. Chem. 282 (2007) 1170–1174.
[171] A. Raturi, T. Simmen, Where the endoplasmic reticulum and the mitochondrion
tie the knot: the mitochondria-associated membrane (MAM), Biochim. Biophys.
Acta 1883 (2012) 213–224.
[172] A. Zhang, C.D. Williamson, D.S. Wong, M.D. Bullough, K.J. Brown, Y. Hathout, A.M.
Colberg-Poley, Quantitative proteomic analyses of human cytomegalovirusinduced restructuring of endoplasmic reticulum–mitochondrial contacts at late
times of infection, Mol. Cell. Proteomics 10 (2011), (M111.009936).
[173] E. Area-Gomez, A.J.C. de Groof, I. Boldogh, T.D. Bird, G.E. Gibson, C.M. Koehler,
W.H. Yu, K.E. Duff, M.P. Yaffe, L.A. Pon, E.A. Schon, Presenilins are enriched in
endoplasmic reticulum membranes associated with mitochondria, Am. J. Pathol.
175 (2009) 1810–1816.
[174] H.J. Wang, G. Guay, L. Pogan, R. Sauvé, I.R. Nabi, Calcium regulates the association
between mitochondria and a smooth subdomain of the endoplasmic reticulum,
J. Cell Biol. 150 (2000) 1489–1498.
[175] T. Hayashi, T.-P. Su, Sigma-1 receptor chaperones at the ER–mitochondrion
interface regulate Ca(2 +) signaling and cell survival, Cell 131 (2007)
596–610.
[176] S.Y. Gilady, M. Bui, E.M. Lynes, M.D. Benson, R. Watts, J.E. Vance, T. Simmen,
Ero1alpha requires oxidizing and normoxic conditions to localize to the
mitochondria-associated membrane (MAM), Cell Stress Chaperones 15 (2010)
619–629.
[177] P. Cosson, A. Marchetti, M. Ravazzola, L. Orci, Mitofusin-2 independent juxtaposition of endoplasmic reticulum and mitochondria: an ultrastructural study,
PLoS One 7 (2012) e46293.
[178] K. Xu, H.P. Babcock, X. Zhuang, Dual-objective STORM reveals three-dimensional
filament organization in the actin cytoskeleton, Nat. Methods 9 (2012) 185–188.
[179] B. Huang, S.A. Jones, B. Brandenburg, X. Zhuang, Whole-cell 3D STORM reveals
interactions between cellular structures with nanometer-scale resolution, Nat.
Methods 5 (2008) 1047–1052.
[180] D. Jiang, L. Zhao, D.E. Clapham, Genome-wide RNAi screen identifies Letm1 as a
mitochondrial Ca2+/H+ antiporter, Science 326 (2009) 144–147.
[181] E. Froschauer, K. Nowikovsky, R.J. Schweyen, Electroneutral K+/H + exchange
in mitochondrial membrane vesicles involves Yol027/Letm1 proteins, Biochim.
Biophys. Acta 1711 (2005) 41–48.
[182] K.S. Dimmer, F. Navoni, A. Casarin, E. Trevisson, S. Endele, A. Winterpacht, L.
Salviati, L. Scorrano, LETM1, deleted in Wolf–Hirschhorn syndrome is required
for normal mitochondrial morphology and cellular viability, Hum. Mol. Genet.
17 (2008) 201–214.
Please cite this article as: S.C.J. Helle, et al., Organization and function of membrane contact sites, Biochim. Biophys. Acta (2013), http://dx.doi.org/
10.1016/j.bbamcr.2013.01.028