Week 8 Linkage 1. whether in coupling or repulsion, recombination
advertisement

Week 8
Linkage
1. whether in coupling or repulsion, recombination fraction is the same
2. reciprocal genotype are always expected to be approximately equal in frequency
unless one of the classes is lethal for some reason
3. 100 % cross over in an interval= 50% recombination =50 M.U. = 1 chiasmata
% recombination(M.U.) = (# of recombinants/total)* 100
[T95]
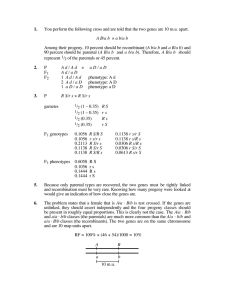
3 genes
-corn
a/a ; s+/s+ ; r+/r+ x a+/a+ ; s/s ; r/r
?
F1 a+/a ; s+/s ; r+/r
Test cross F1 a+/a ; s+/s ; r+/r x
a/a ; s/s ; r/r
Questions:
Are the 3 genes linked? 2 linked and l unlinked? All 3 linked?
[T 95a]
-test cross results indicate the 3 genes are linked
4 genes
1. all unlinked
2. 3 linked, 1 unlinked
3. 2 unlinked and 2 linked
4. 2 groups of 2 linked genes
5. all 4 linked
For each chromosome represented (linkage groups), 2 relatively abundant non cross over
classes will be produced by independent assortment of genes on separate chromosomes
[T 96]
Mapping calculations
-best genetic mapls are those that involve single cross over
-a way to attempt this is to use as many markers as possible
-if cross over occurs at random position along the chromosome, then the expected double
cross over frequency would be:
(0.41*0.728*2501)/100=7.5(frequency of double cross over )
observed # based on [T 95a]=2 , expected =7.5
-this indicate interference
[T 97]
-recombinant fraction
1 (2 strand double cross over) : 2 (3 strand double cross over) : 1 (4 strand double cross
over)
Nature of crossing over
-1931, Creighton & McClintock
-they demonstrated that cross over and genetic recombination involves the physical
exchange of parts of homologous chromosomes
-they also demonstrated that crossing over occurs after S-phase
(observed 50 % recombinants and 50% parental)
-but the decision had to be made for organism where it’s pausible to recover and analyze
all 4 products of a mieotic tetrad
Attached X in D. mel.
-can see 4 products of mieotic event
e.g.
female : (Xv+Xv ) attached Y x Xw+ Y
v=vermillion
-if cross over is pre-S , all progeny are v+
-if cross over is post-S, see vermillion progeny
Is there cross over between sister chromatids?
-yes
[97a]
-chiasmata
-1978
Tease and Jones proved that there are cross over between sister chromatids
-they examined Harlequin chromosomes (stained such that homologs have different
colors)
-let chromosomes replicate thru. 2 rounds in Budr (Bromodeoxyuridine)
-then stain the chromosome with Giemca and fluorescent dye
-then examine under microsope
-the chromatids with 2 Budr helice stain lightly and the chromatid with 1 Budr stain
darker
[T 98], [T 99]
-shows sister chromatid exchange
Fungal Genetics
Ascus (bag) containing all products of a single meiosis
-so the isolation of asic allows one to examine the genotypes of all the product from a
single meiocyte
[T 100]
Mapping centromeres
-phenotype of centromere is lst division segregation
[T101]
Map distance for gene A and the centromere
{0.5(# of 2nd division)/total}*100
[T 102, 103, 103a]
Mapping between genes
-how do we detect linkage?
-if NPD= PD, indicate that the 2 genes are unlinked
[T 104, 105, 106]
-frequency of tetratype is determined by the distance of the respective gene from the
centromere
-if gene is further from the centromere, frequency of tetratype increases
If the 2 genes are linked, PD>> NPD
-therefore genes are staying together in parental combinations
-no crossover= PD and FDS for both genes but a crossover between b and the centromere
will also produce a PD whe re both genes now show SDS (2:2:2:2)
-cross over between a and b, either SDS for a or b
-4 chromatid double cross over gives NPD
[T 107]
What is the map distance between a and b for these data?
Recombinat frequency (r.f.) = # recombinants/total
r.f. = 50(TT + 6NPD)/total
-this formula considers double cross overs
simple formula = 0.5(TT) + NPD/total
[T 107]
r.f. = 50(26)+3/100
r.f. = 16 M.U.
however with the formula that take double cross overs into consideration:
r.f.= 50(26+6*3)/100
r.f.= 22 M.U.
Why r.f.= 50(TT+6 NPD)/total
[T 108, 109]
[T 110]
with a map, you can deduce where the cross over must occur to produce a particular
spore genotype arrangement
[T 110, 111, 112, 113]
-are the genes linked?
-if yes, what is the map?
-if no, what are the gene, centromere distances?