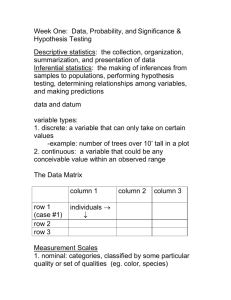
BIOL300 Lab Manual 2014S
advertisement

BIOL 300 Spring 2014 Lab Manual for the Analysis of Biological Data © 2008-2013 Michael Whitlock. Chapter 2 © Michael Whitlock, Brad Anholt, and Darren Irwin Biology 300 lab manual, Introduction This book contains instructions for the lab exercises for BIOL300, the biostatistics course at UBC. It is intended to complement, not to replace, the text Analysis of Biological Data, by Whitlock and Schluter. The labs contain a mix of data collection, computer simulation, and analysis of data using a computer program called JMP (pronounced "jump"). All data described in these labs are real, taken from actual experiments and observations reported in the scientific literature. References for each paper are given in at the end of the manual. This lab manual is a work-in-progress. Suggestions for improvements are welcome. This manual has been much improved by suggestions from Gwylim Blackburn, Becca Gooding-Graf, Darren Irwin, Haley Kenyon, Heather Kharouba, Becca Kordas, and Jess McKenzie. Many of the labs use java applets, and these are to be found on the web. In order to save you from typing those URLs, links are provided from the lab page at http://www.zoology.ubc.ca/~irwin/BIOL300/labs/. 2 Biology 300 lab manual, Lab 1 1. Introduction to statistics on the computer and graphics Goals Get started on the computers, learning how to start with JMP Collect a data set on ourselves for future use Make graphs, such as histograms, bar charts, box plots, scatter plots, dot plots, and mosaic plots. Learn to graph data as the first step in data analysis Quick summary from text (see Chapter 2 in Whitlock and Schluter) Computers make data analysis faster and easier. However, it is still the human's job to choose the right procedures. Graphing data is an essential step in data analysis and presentation. The human mind receives information much better visually than verbally or mathematically. Variables are either numerical (measured as numbers) or categorical (describing which category an individual belongs to). The distribution of categorical variables can be presented in a bar chart. The distribution of numerical variables can be presented in a histogram, a box plot, or cumulative frequency plot. The relationship between two numerical variables can be shown in a scatter plot. The relationship between two categorical variables can be shown in a mosaic plot or a grouped bar chart. The association between a numerical variable and a categorical variable can be shown with multiple histograms, grouped cumulative frequency plots, or multiple box plots. A good graph should be honest and easy to interpret, with as much information as needed to interpret the graph readily available. At the same time, the graph should be uncluttered and clear. 3 Biology 300 lab manual, Lab 1 Activities 1. Log on to the computer with your new password. (See the next section if you need detailed instructions.) Remember this username and password; you'll need them throughout the term. 2. Recording data. Using the "Student data sheet 1" at the end of this manual, record the requested information about yourself. This is optional; if you have any reason to not want to record this (relatively innocuous) data about yourself, you do not have to. If you feel that you would like to skip just one of the bits of information and fill in the rest, that is fine too. The data sheets do not identify students by name. Pass the sheet to the instructor when you are finished. Learning the Tools 1. How to log on a. If the screen is dark, make sure the screen is turned on, with the lighted button in the bottom right corner. Move the mouse or hit the shift key to wake up the computer. b. You may be prompted to hit Ctrl-Alt-Delete (three buttons all at the same time). Hit OK to the next screen. c. Enter the username and password into the appropriate places, from the slip of paper given to you by the TA. Keep track of these; you'll be using these throughout the term. d. After the computer starts up, double-click on the icon for "JMP". This is the statistical package we'll use most in this class. e. After you're done for the day, don’t forget to log-off using the menu in the lower left corner of the screen and hit OK. 2. Opening a data file Using the JMP starter, click on "Open data table." This lets you open previously saved files. Navigate to the shared drive called "Shared on 'Statslab Server (Statslab)'". This is where all the previously collected data sets needed for this class are stored. You can also create and store new data sets or copies of these others in your own folders with your account. 4 Biology 300 lab manual, Lab 1 Open a file called "titanic.csv". This file has information on all the passengers of the RMS Titanic on its initial and disastrous run. 3. Check that the variables are labeled as “continuous” or “nominal” correctly. The and icons that you see to the left of your data file tell you what the computer thinks is the type of each of your variable. The icon denotes a numerical variable (what JMP calls “Continuous”), and the icon makes categorical variables (what JMP calls “Nominal”). If the type is incorrect, just click on the or from the menu that appears. icon and choose the correct type 4. Changing column information. You can change information about the column (e.g. its title, the data type, etc.) by double-clicking on the column heading. A straightforward menu will appear. 5. Making a graph of one variable For example, using the Titanic data in "titanic.csv", let's plot a histogram of the ages of the passengers. From the menu bar, click "Analyze" and under that menu click "Distribution". A window will appear. In that window click on "Age" under "Select columns", then click on the button labeled "Y, Distributions" under the second column, and then click OK. 5 Biology 300 lab manual, Lab 1 A window will appear that includes the histogram, although it will be turned on its side. (If you want the histogram to appear in the more typical horizontal layout, click on the red triangle next to the variable name "Age" to open a menu, click "Display Options" and then "Horizontal Layout".) We can plot a bar chart showing the distribution of a categorical variable in exactly the same way, for example with the variable "Passenger class." (Note: JMP makes bar charts without spaces between the bars, which is a flaw in the program.) 6. Making a graph of two or more variables To plot the relationship between two variables, choose "Fit Y by X" from the "Analyze" menu bar listing. In the window that opens, select the explanatory variable you are interested in from the list on the left, and then click the button labeled "X, Factor" from the middle column. For example, click on "Sex" and then click "X, Factor." Then click on the response variable from the list on the left, and then click the button labeled "Y, Response". Finally click "OK". A window will appear that shows an appropriate graph of the relationship between the two variables. JMP will choose a graphic type that matches the variable types of the selected variables, so the same procedure will give a mosaic plot for two categorical variables, a scatter plot for two numerical variables, or dot plots for one categorical and one numerical variable. 6 Biology 300 lab manual, Lab 1 7. Making graphs for more than one group at a time. In either the "Fit Y by X" or "Distribution" menus, you can plot graphs of subsets of the data by clicking on one of the variables and clicking the "By" button. For example, to plot histograms of height for each sex separately, put "Age" in the "Y, Distribution" box of the "Distribution" window, and put "Sex" in the "By" box. JMP will give you separate results for males and females. 8. Using the manual. These course notes will only scratch the surface of the possible techniques available in JMP. You can find out about more features by using the online manuals. The search function on JMP help is substandard, so you may find it easiest to look in the index of one of the books available online, such as the Statistics and Graphics Guide. Look under the "Help" menu for "Books" and choose the "JMP Stat and Graph Guide". 9. JMP tips At the end of this lab manual, there are a couple of pages with some general tips on how to use JMP more easily and powerfully. 10. Box plots JMP automatically graphs numerical data using box plots, typically shown to the right of the histogram. Below is the box plot for the variable "Mortality rate adult female" from the "countries2005.csv" file. You will learn about the various things the box plot shows later (e.g. lab 3), but we show it now to increase familiarity with the graphing output of JMP. Essentially, the box plot illustrates at a glance where the bulk of the distribution is (in the box) and the range of the data, as well as other details. We've added labels of the various parts: 7 Biology 300 lab manual, Lab 1 Questions 1. For each of the following pairs of graphs, identify features that are better on one version that the other. a. Survivorship as a function of sex for passengers of the RMS Titanic b. Ear length in male humans as a function of age 2. Open the data file called "countries2005." This file gives various data from the World Bank on all countries of the world for 2005. a. Plot a histogram for the variable "birth rate crude (per 1000 people)." b. Look carefully at the histogram. Do you see evidence of a problem with the data file? Find and fix the problem. (Two practical hints: 1. An individual can be identified from a plot by clicking on a dot or bar of the histogram. This will select the row or rows associated with that part of the graph. 2. A row can be excluded from analysis by selecting the row and then choosing "Delete Rows" from the "Rows" menu.) c. Plot the histogram on the corrected data set. 8 Biology 300 lab manual, Lab 1 3. Muchala (2006) measured the length of the tongues of eleven different species of South American bats, as well as the length of their palates (to get an indication of the size of their mouths). All of these bats feed on nectar from flowers, using their tongues. Data from their paper are given in the file "battongues.csv." a. Plot a scatter plot with palate length as the explanatory variable and tongue length as the response variable. Describe what you see. b. All of the data points in this graph have been double checked and verified. With that in mind, how do you interpret the outlier on the scatter plot? (You might want to read about these bats in the Muchhala (2006) paper, to learn more about the biology behind Anoura fistulata.) 4. Use the countries data (as corrected as per Question 2). Plot distributions for "Continent," "Prevalence of HIV," and "Physicians per 1000 people." What kinds of variables are these (numerical or categorical)? What kinds of graphs are being drawn? 5. Use the countries data (as corrected in Question 2). Plot the relationship between the following sets of variables: a. Male life expectancy and female life expectancy, b. Continent and life expectancy, c. Literacy rates and life expectancy d. Personal computers and life expectancy e. Number of physicians and life expectancy f. Which variable seems to explain life expectancy better: number of personal computers or number of physicians? Try to explain the pattern that you see. 9 Biology 300 lab manual, Lab 1 6. Use the data set collected on your class from today. (We'll return later to some of the other variables later in the term.) Plot the relationship between the following pairs of variables. For each case describe the pattern that you observe: a. Handedness and "footedness" b. Handedness and dominant eye c. Sex and height d. Height and head circumference 7. Use the data from class today. Plot the distributions of height for males and females separately. 8. Pick one of the plots made by the computer program. What could be improved about this graph to make it a more effective presentation of the data? 9. (Optional) -- This last exercise will teach some slightly more advanced uses techniques for using JMP. a. JMP will do other kinds of plot as well. For example, for the distribution of a numerical variable, you can plot Cumulative Frequency Distributions (CDF) or stem-and-leaf plots. To do so, get to the results window of the Distribution process, and then under the red triangle click on "CDF Plot" or "Stem and Leaf." b. Many features on JMP are quite intuitive, especially once you know about the tiny red triangles . Play around with the program and a data set to see what you can discover. 10 Biology 300 lab manual, Lab 2 2. Sampling Goals Explore the importance of random sampling Investigate sampling error and sampling bias Learn to create data files. Quick summary from text (Chpt. 1 in Whitlock and Schluter) With random sampling, all individuals in the population have an equal and independent chance of being selected for the sample. Most estimation methods and hypothesis tests assume that the data are a random sample from the population. Sampling error is the chance difference between an estimate describing a sample and that parameter of the whole population. (Note that sampling error is not due to measurement error or sampling bias, but rather arises naturally from the process of random sampling.) If individuals are not taken independently of each other, then the sample is effectively smaller than it seems. The resulting estimates will have higher sampling error than those taken from the same size random sample. If some individuals are more likely than others to be selected for the sample, then the sample has a high probability of being biased. If so, we say that the process is subject to sampling bias. Estimates are made from data. The goal of an estimate is to give a close idea of the true value of a parameter in a population. Estimates are almost always wrong, in the sense that they rarely exactly match the true value of the parameter. Estimates can be biased, meaning that on average multiple estimate taken in the same way will be different from the true value. Estimates can also be imprecise, meaning that different samples will give different estimated values. 11 Biology 300 lab manual, Lab 2 Learning the tools In most computer programs for data analysis, data is entered in a particular way into a table, or spreadsheet. These spreadsheets are divided into lots of small rectangles called cells, and each cell belongs to a column (a vertical stack of cells) and a row (a horizontal set of cells). The convention is that each row corresponds to all the data from a single individual, and each column gives the data for all individuals for a single variable. Key point: In a stats spreadsheet like JMP, each row is a distinct individual, and each column is a variable. For example, here is a part of a data file on the passengers of the Titanic, which shows the rows for 10 individuals, and for each individual, their values for seven different variable: passenger class, name, age, the place they embarked from, their destination, their sex, and whether they lived or died during the disaster, with a yes or no. The full data set includes many more rows, for all the other passengers aboard the ship. In the first row, for example, we see that a female passenger named Miss Elisabeth Walton Allen traveled in first class from Southampton to St. Louis, and she survived the journey. Setting up a new data file is an extremely important part of data analysis. If variables are not labeled clearly and unambiguously, the results will not be interpretable later. 1. Open a new file. In JMP, click on the "New Data Table" button from the JMP starter. Save it as "disaster.jmp." (Note -- we're not going to use this new file; this is just for an example of how to do the steps.) 2. Make new columns. For each variable in your data, add a new column. Under the menu labeled "Cols" choose "Add multiple columns." Enter the number of variables in the data, in this case 7, to the box labeled "How many columns to add." Click "OK." 12 Biology 300 lab manual, Lab 2 For each column, change the label to the desired variable name and tell the computer the type of variable it is. For example, for the first column, double click on the white space at the top of the column. A new window will open which allows you edit the column name (put "Passenger Class"). Also, this data is not numerical but is based on alphabetic characters. So click on the "Data type" window and click on "character." "Modeling type" should also change to "nominal" as a result. These changes tell the computer to treat the data in this column as categorical data, not as numerical data. This is an essential step in having the analysis work correctly later. Finish this with other columns, if you like. Name, embarked, destination, sex, and survived are categorical variables, while age is a numerical variable. 3. Enter the data. For each individual, add a row and type in the values for that individual into the appropriate cells. The "Tab" key will take you easily to the next cell. JMP will automatically start a new row if you hit "Tab" in the last cell of the previous row. 4. Summarizing numerical data To find the mean of a numerical variable, choose "Distribution" from the "Analyze" menu. Click on the variable name you are interested in (for example Age), then click on "Y, columns" and OK. The mean of the variable will appear among a list of summaries of the data below the graph. 13 Biology 300 lab manual, Lab 2 5. Creating new columns using other variables. Sometimes it can be useful to create new variables that use other data already in the data file. For example, we might want to convert a value measured in inches to one measured in centimeters. We might want to take the log of each data point. Or, as we will need later, we might want to calculate the difference between two variables. As an example, let's make a column with approximate age in months. To calculate the new variable, first create a new column in the same way as above ("Cols" > "New Column..."). Name the new variable in the resulting box, and choose the correct data type for the new variable. In this case, let's make a numerical variable called "age (months)". After returning to the data window, click on any cell in the new column, and then click on "Formula..." under the "Cols" menu. A new window will open. This window contains a space at the bottom to create a formula that uses other data and other mathematical functions. To make our new column, double click on "Age" to add it to the bottom window, then click on the multiplication sign, then enter 12 to the new box and hit return. When finished building the formula, hit the "OK" button. 14 Biology 300 lab manual, Lab 2 Activities In this class you will be measuring several cowrie shells and then comparing the measurements by using the computer to plot the data. The class divides itself into groups of two to four people. Each group will be given a container of cowrie shells and a white board. Follow these steps: 1. As a group, sample five shells from this container. Put them in spots 1 through 5 on the white board. This is your sample. 2. For each shell that you have sampled, measure the length of the shell, and write these lengths down on the white board by the shell. 3. Open a new data file in JMP, and enter the lengths of your five shells. Save this with a title something like “First sample.jmp”. 4. Calculate the mean and standard deviation of the length of these five shells. Record this mean. 5. Next we will calculate the mean of the population of shells in your container. Take each remaining shell one by one, measure its length, and record it on the whiteboard beside the next available number. Set the shell there on the whiteboard as well. 6. Open another data file in JMP. Add the lengths of all the shells into this file under a column for length. Save this as something like “Full population shells.jmp”. 7. Calculate the mean and standard deviation of the full population. 8. Is the mean of your sample the same as the mean of the population as a whole? What about the standard deviation? Explain reasons why the sample mean and the population mean could differ. 9. Let’s now explore how to randomly sample from the population. You should have each shell now next to a unique number on your whiteboard. To randomly choose five of these for a random sample, we can use random numbers generated by http://www.random.org/sequences/. Open this web page, tell it that the “smallest value” is 1 and the “largest value” is the number of shells in your population. Click “Get sequence”. Use the first five numbers in the list to tell you which shells to sample. 10. We’re going to randomly sample from the population several times. Each sample is going to have five shells, and we’ll calculate the mean of each sample. Open a new file in JMP, and give it two columns. The first column will tell us which sample an individual belongs to, and this should be specified as a nominal 15 Biology 300 lab manual, Lab 2 (categorical) variable called “Sample”. The second column will be the length measurement for a randomly chosen shell, so this column should be specified as a “continuous” (i.e. numerical) variable, called “Length.” 11. For the first sample, put “S1” in each of the first five rows of this file in the “Sample” column. “S1” will indicate that an individual was part of “sample 1.” This sample will have five individuals in it, so we need five rows. For the second sample, you’ll want five rows with “S2” for the label of the sample, etc., in the same file. 12. Record the length of the five shells in your first random sample, in the second column, one length for each row. 13. Repeat steps 11 and 12 thirty times, changing the “Sample” variable appropriately (“S1” through “S30”). Ask the random number generator for a new set of random numbers each time to choose which shells to use for each sample. 14. Create a new file that has the mean shell lengths for each of the randomly chosen samples. You can do this with the “Tables” menu bar, and then select “Summary”. In the resulting window, put the name of your sample label column in the box for Groups. The select the name of the length column, and then click “Mean” in the pop-up menu that appears when you click “Statistics”. When you hit “OK”, JMP should create a new data file with the means of each sample. Questions 1. Plot a histogram in JMP that shows the distribution of the means of your random samples. Describe the shape of this distribution. Does every sample return the same value of the mean? Why, or why not? 16 Biology 300 lab manual, Lab 2 2. What is the mean of the distribution of the random sample means? Is it close to the mean shell length of the population? 3. How does the mean of your very first sample (in step 1) compare to the population mean? Is it more or less different from the population mean than a typical sample mean from the randomized samples? If so, why might that be? Assignment (not to be done in Spring 2014, but good to think about anyway) Write a report documenting the results of your sampling exercises this week. Address these two questions: "Is there evidence of sampling bias in your fist sample?" and " How much does sampling error affect the reliability of using a sample of five individuals to estimate the mean shell length of a population?" Write the report in the format of a scientific paper, with a brief introduction describing the questions, a methods section (which needs to specify your sampling system as well as the basic approach detailed above), results, and discussion. The report should not require more than 3-4 pages of double-spaced typed text. More advice on writing a good report is given at the end of this lab manual. 17 Biology 300 lab manual, Lab 3 3. Describing data and estimation Goals Investigate sampling error; see that larger samples have less sampling error. Visualize confidence intervals. Calculate basic summary statistics using the computer. Calculate confidence intervals for the mean on the computer. Understand box plots. Quick summary from text (Chpts. 3 and 4 in Whitlock and Schluter) The mean describes the "center of gravity" of a set of numbers. The median is the data point that is smaller than half of the data and greater than the other half of the data. The standard deviation is a description of the typical spread of data around the mean. The standard deviation is the square root of the variance, which is the average squared deviation from the mean. The 3rd quartile is the data point greater than three quarters of the other data. The 1st quartile is the point greater than one-quarter of the other data. The interquartile range is the difference between the third and first quartiles. The interquartile range is another measure of the spread of a set of data. The skew of a set of data describes the asymmetry of the distribution. Zero skew means that the distribution is symmetric. If the skewness is positive, then the distribution tends to have a long tail to the right. With negative skewness, the distribution has a long tail to the left. Sampling error is the variation in estimates among samples caused by chance deviations between the sample and population. Sampling error is greater in small samples than in large samples. Large samples are more likely to give a precise estimate. Sampling error is measured by the standard error. The standard error is the standard deviation of the sampling distribution of an estimate. The standard error of a mean can be calculated by the standard deviation divided by the square root of the sample size: SEY = s / n . 18 Biology 300 lab manual, Lab 3 The reliability of an estimate can be expressed as a confidence interval. a 95% confidence interval will contain the true value of the parameter in 95% of estimates. Activities Distribution of sample means Go to the web and open the page at http://onlinestatbook.com/stat_sim/sampling_dist/index.html. This page contains a java applet that lets you play around with sampling, to see the distribution of sampling means. After the page loads, click the "Begin" button. You'll see a window that looks like this: The top box contains a distribution that represents the true distribution of a variable in the population. This is the true universe that the program is using to sample individual data points from. In the example pictured here, the mean of the population is 16, with standard deviation 5, and the individuals in the population occur according to a normal (bell-shaped) distribution. Next click the "Animated" button on the right side. This will draw five individuals at random from the population, and plot a histogram of the sample data in the second graph. It will also calculate the mean of the sample, and start to create a histogram in the third graph. The third graph shows the distribution not of individuals, but of sample means. Notice how the bar added in the distribution of sample means corresponds to the mean of the sample in the second graph. If you click "Animated" again, it will draw an entirely new sample from the population, and then add the mean of that new sample to the distribution of sample means. Do this a few times, and watch what's happening. 19 Biology 300 lab manual, Lab 3 After you get bored of that, click the button that says "5". This button will draw 5 different samples, and add the five new sample means to the histogram of sample means. The "1,000" and "10,000" buttons do the same thing, except for more samples. After adding a large number of samples, look at the distribution of sample means. What kind of shape does it have? It ought to look normal as well. Also look at the width of the distribution of sample means - is it more variable, less variable or about as variable as the population distribution? What do you expect? Next, hit the "Clear lower 3" button at the top. This will erase all the distributions that you just drew. Use the "N=5" button to choose a new sample size, starting with N=25. Repeat the steps above to generate a distribution of sample means. Now the sample means are based on 25 individuals instead of just 5 individuals. Are the sample means more or less variable with N=25 than with N=5? Confidence intervals Go back to the web, and open the java applet at http://onlinestatbook.com/stat_sim/conf_interval/index.html. This applet draws 100 different confidence intervals for the mean of a known population. Hit "Sample," and look at the results. Each horizontal line on the graph represents a different sample of 10 individuals (or more, if you change the sample size). Each line shows the 95% confidence interval for the mean (the inside part of the line in red or yellow) as well as the 99% confidence interval (the wider line in blue or white). The 95% confidence interval is shown in yellow if the interval contains the true value of the mean, represented by the vertical line down the screen. The program changes the 95% confidence interval line to red if the 95% confidence interval fails to enclose to true mean. Out of the 100 samples, how many 95% confidence intervals contain the true mean? How many missed? Click "Sample" a few more times. Each time you click, the program will draw 100 more samples and draw them. It also keeps track of all the sample you have drawn in the table at the bottom. After drawing a lot of samples (say 1000), what proportion of samples have 95% confidence intervals that include the true mean? What proportion fail to enclose the true mean? What about the 99% confidence interval -- what fraction of 99% confidence intervals enclose the true mean? A 95% confidence interval ought to enclose the true value of the parameter for 95 samples out of 100, on average, and the 99% confidence interval ought to succeed 99% of samples. In fact, of course, that is the definition of a 95% (or 99%) confidence interval; the percentage tells us the expected probability that the true mean is captured within the confidence interval. 20 Biology 300 lab manual, Lab 3 Learning the Tools 1. Calculating summary statistics a. Use the "Distribution" window, found under the "Analyze" menu bar. In the window that opens, choose the variable (or variables -- you can do more than one variable at once), click "Y-columns", and hit "OK." A graph will appear, and below the graph will be a collection of summary statistics including mean, standard deviation, standard error of the mean, and the 95% confidence interval for the mean. b. To find a longer list of summary statistics, click on the red triangle next to the variable name. From the resulting menu, click on "Display Options" and then "More moments." This will add to the list of summary statistics, including the variance, skewness, and the coefficient of variation (CV). Key point: The red triangles sprinkled through JMP windows open new menus of options when clicked. 2. Calculating confidence intervals of means The 95% confidence interval is automatically calculated by opening the "Distribution" window for a variable. For other confidence intervals, open the "Distribution" window, and the click the red triangle next to the variable name. Select "Confidence Intervals" and choose the desired confidence level from the list. (For example, for a 99% confidence interval, choose "0.99"). This will add a section to the results with the confidence intervals for the mean and standard deviation of the variable. 21 Biology 300 lab manual, Lab 3 Questions 1. The data file "students.csv" should include data that your class collected on themselves during the first week of lab, in chapter 1. Open that file. a. What is the mean height of all students in the class? b. Look at the distribution of heights in the class. Describe the shape of the distribution. Is it symmetric or skewed? Is it unimodal or bimodal? c. Calculate the standard deviation of height. d. Calculate the standard error of height. Does this match the SEY = s / n that you expect? e. Calculate the mean height separately for each sex. Which is higher? Compare the standard deviations of the two sexes as well. 2. The file "caffeine.csv" contains data on the amount of caffeine in a 16 oz. cup of coffee obtained from various vendors. For context, doses of caffeine over 25 mg are enough to increase anxiety in some people, and doses over 300 to 360 mg are enough to significantly increase heart rate in most people. Red Bull contains 80mg of caffeine per serving. a. What is the mean amount of caffeine in 16 oz. coffees? b. What is the 95% confidence interval for the mean? c. Plot the distribution of caffeine level for these data. Is the amount of caffeine relatively predictable in a cup of coffee? What is the standard deviation of caffeine level? What is the coefficient of variation? d. The file "caffeine-Starbucks.csv" has data on six 16 oz. cups of coffee sampled on six different days from a Starbucks location, for the Breakfast Blend variety. Calculate the mean (and its 95% confidence interval) for these data. Compare these results to the data taken on the broader sample in the first file, and describe the difference. 3. A confidence interval gives a range of values that are likely to contain the true value for a parameter. Consider the "caffeine.csv" data again. 22 Biology 300 lab manual, Lab 3 a. Calculate the 95% confidence interval for the mean caffeine level. b. Calculate the 99% confidence interval for the mean caffeine level. c. Which is larger (i.e. has a broader interval)? Why should this one be larger? d. Calculate the quantiles of the distribution of caffeine. (Big hint: they automatically appear in the "Distribution " window, immediately below the graphs.) What are the 2.5% and 97.5% quantiles of the distribution of caffeine? Are these the same as the boundaries of the 95% confidence interval? If not, why not? Which should bound a smaller region, the quantile or the confidence interval of the mean? 4. Return to the class data set, "students.csv." Find the mean value of "number of siblings." Add one to this to find the mean number of children per family in the class. a. The mean number of offspring per family twenty years ago was about 2. Is the value for this class similar, greater, or smaller? If different, think of reasons for the difference. b. Are the families represented in this class systematically different from the population at large? Is there a potential sampling bias? c. Consider the way in which the data was collected. How many families with zero children are represented? Why? What effect does this have on the estimated mean family size? 5. Return to the data on countries of the world, in "countries2005.csv" and the data from the class in "students.csv." Plot the distributions and calculate summary statistics for land area, life expectancy (total), personal computers per 100 people, physicians (per 1000 people), as well as height for each sex. Key point: A distribution is skewed if it is asymmetric. A distribution is skewed right if there is a long tail to the right, and skewed left if there is a long tail to the left. a. For each variable, plot a histogram of the distribution. Is the variable skewed? If so, in which direction? 23 Biology 300 lab manual, Lab 3 b. For each variable, calculate the mean and median. Are the similar? Match the difference in mean and median to the direction of skew on the histogram. Do you see a pattern? 6. A box plot is a graphical technique to show some key features of the distribution of a numerical variable. Unfortunately, box plots are not always drawn in exactly the same way. JMP draws box plots automatically with the "Distribution" window, to the right of the histogram. Plot the box plot for "Mortality rate adult female" from the "countries2005.csv" file. It should look like this, except that we've added labels of the various parts: The interquartile range is the difference between the 75th percentile and the 25th percentile. It is an alternate measure of the spread of the distribution. Look at the values in the Quantiles and Moments sections below the graphs, and read them off this graph. Confirm that these designations on this graph are correct. 24 Biology 300 lab manual, Lab 4 4. Probability Goals Practice making probability calculations Investigate probability distributions Quick summary from text (Chpt 5 in Whitlock and Schluter) The addition principle says that the probability of either of two mutually exclusive events is the probability of the first event plus the probability of the second event: Pr[A or B] = Pr[A] + Pr[B] . The general addition principle says that the probability of either of two events is the probability of the first event plus the probability of the second event, minus the probability of getting both events: Pr[A or B] = Pr[A] + Pr[B] − Pr[A and B]. The multiplication principle says that the probability of two events both occurring - if the two events are independent -- is the probability of the first times the probability of the second: Pr[A and B] = Pr[A] Pr[B]. The general multiplication rule says that the probability of two events both occurring is the probability of the first event times the probability of the second event give the first: Pr[A and B] = Pr[A] Pr[B | A]. A probability tree is a graphical device for calculating the probabilities of combinations of events. The law of total probability, Pr [ A ] = ∑ Pr [ B ] Pr [ A | B ] , makes it possible to all B calculate the probability of an event (A) from all of the conditional probabilities of that event. The law adds up for all possible conditions (B) the probability of that condition (Pr[B]) times the conditional probability of the event assuming that condition (Pr[A | B]). The binomial distribution describes the probability of a given number of successes out of n trials, where each trial independently has a p ! n $ X n−X probability of success: Pr [ X ] = # & p (1− p) . " X % 25 Biology 300 lab manual, Lab 4 Activities 1. Let's Make a Deal An old TV game show called "Let's Make a Deal" featured a host, Monty Hall, who would offer various deals to members of the audience. In one such game, he presented a player with three doors. Behind one door was a fabulous prize (say, a giant pile of money), but behind the other two were relatively worthless gag gifts (say, donkeys). The player was invited to choose one of the doors. Afterwards, the host would open one of the other two doors to reveal that it contained a donkey. Then, the player is offered a choice: they can either stay with their original door choice, or they can switch to the other closed door. The question becomes, which strategy is better -- to stick with the original or to switch doors? In the last few years, this relatively simple question has attracted a lot of debate, with professional mathematicians getting different answers. a. Find the probability of finding the door with the pile of money, if the player follows the strategy of staying with their original choice. b. Find the probability of success with strategy of switching to the other closed door. c. Using the simulation at http://www.stayorswitch.com, play the game 20-30 times using each of the two strategies. Was your answer correct? Remember that because the game in the simulation is a random process, the proportion of successes will not necessarily exactly match your theoretical predictions. (Note that there is a version of the game at another website that will keep track of your results, although it is a little clunky to use: http://www.stat.tamu.edu/~west/applets/LetsMakeaDeal.html 2. The binomial distribution This is a simple, quick exercise meant to help you visualize the meaning of a binomial distribution. The binomial distribution applies to cases where we do a set number of trials, and for each trial there is an equal and independent probability of a success. Let's say there are n trials and the probability of success is p. Of those n trials, anywhere between 0 and n of them could be a 'success" and the rest will be "failures." Let's call the number of successes X. This is relevant to many biological situations; for example, calculating the probability of getting exactly three daughters in a family of five, when you assume a certain fixed probability of each baby being female. 26 Biology 300 lab manual, Lab 4 As our example, let's roll five regular dice, and keep track of how many come up as "three." In this case, there are five trials, so n = 5. The probability of rolling a "three" is 1/6, so p = 0.1666. Let's repeat the sampling process a lot of times. For each sample, roll five dice, and record the number of threes. For this exercise, let’s draw a histogram of the results by hand. First draw the axes, and then for each result draw an X stacked up in the appropriate place. For example, after three trials that got 0, 1, and 1 threes, respectively, the histogram will look like this: Keep doing this until you have 20 or more samples. Here is the frequency distribution for this process as calculated from the binomial distribution. Is this similar to what you found? (Remember that with only a few trials, you don’t expect to exactly match the predicted distribution. If you increase your number of samples, the fit should improve.) The expected mean number of threes is 0.833. (For the binomial distribution, the mean is n p, or in this case 5 × 0.1666.) Calculate the mean number of threes in your samples. 27 Biology 300 lab manual, Lab 5 5. Frequency data and proportions Goals Better understand the logic of hypothesis testing. Make hypothesis tests and make estimates about proportions. Fit frequency data to a model. Quick summary from text (Chpts 7 + 8 from Whitlock and Schluter) The null hypothesis (H0) is a specific claim about a parameter. The null hypothesis is the default hypothesis, the one assumed to be true unless the data lead us to reject it. A good null hypothesis would be interesting to reject. The alternative hypothesis (HA) includes all values for the parameter other than that stated in the null hypothesis. The confidence interval for a proportion is best calculated by the AgrestiCoull method. Most statistical packages on the computer do not calculate this method. A binomial test is a hypothesis test that compares a hypothesized value of a proportion to count data. Categorical data describes which category an individual belongs to. Categorical data can be summarized by the frequency or count of individuals belonging to each category. In some cases, the frequency of each category can be predicted by a null hypothesis. We can test such null hypotheses with Goodness-of-Fit tests. A common Goodness-of-Fit test is the χ2 test, which compares the number of individuals observed in each category to the number expected by a null hypothesis. The χ2 Goodness-of-Fit test assumes that each individual is sampled randomly and independently. The χ2 test is an approximate test that requires that the expected numbers in each category is not less than one, and that no more than 20% of the categories have expected values less than 5. 28 Biology 300 lab manual, Lab 5 The χ2 test can be used as a quick approximation to the binomial test, provided that the expected values of both categories are 5 or greater. The Poisson distribution predicts the number of successes per unit time or space, assuming that all successes are independent of each other and happen with equal probability over space or time. Activity We'll do an experiment on ourselves. The point of the experiment needs to remain obscure until after the data is collected, so as to not bias the data collection process. After this paragraph, we reprint the last paragraph of Darwin's Origin of Species. Please read through this paragraph, and circle every letter "t". Please proceed at a normal reading speed. If you ever realize that you missed a "t" in a previous word, do not retrace your steps to encircle the "t". You are not expected to get every "t", so don't slow down your reading to get the "t"s. k It is interesting to contemplate an entangled bank, clothed with many plants of many kinds, with birds singing on the bushes, with various insects flitting about, and with worms crawling through the damp earth, and to reflect that these elaborately constructed forms, so different from each other, and dependent on each other in so complex a manner, have all been produced by laws acting around us. These laws, taken in the largest sense, being Growth with Reproduction; inheritance which is almost implied by reproduction; Variability from the indirect and direct action of the external conditions of life, and from use and disuse; a Ratio of Increase so high as to lead to a Struggle for Life, and as a consequence to Natural Selection, entailing Divergence of Character and the Extinction of less-improved forms. Thus, from the war of nature, from famine and death, the most exalted object which we are capable of conceiving, namely, the production of the higher animals, directly follows. There is grandeur in this view of life, with its several powers, having been originally breathed into a few forms or into one; and that, whilst this planet has gone cycling on according to the fixed law of gravity, from so simple a beginning endless forms most beautiful and most wonderful have been, and are being, evolved. k Problem number 6 will return to this exercise. Please don't read Problem 5 until you have completed this procedure. Learning the Tools 1. Binomial tests 29 Biology 300 lab manual, Lab 5 A binomial test compares the proportion of individuals in a sample that have some quality to a hypothesis about that proportion. In JMP, binomial tests can be made from the "Distribution" window. From the menu bar, choose "Analyze", then "Distribution" and then choose the variable that you want to study and hit OK. For example, the data set called "bumpus.csv" contains data on a sparrow population caught in a wind storm in the 1880's. These data were among the first to demonstrate natural selection in a wild population. Use "Distribution" for the "sex" variable. In the Distribution window, choose "Test Probabilities" from the menu that opens from the red triangle by the variable name. This expands the results window below, and give a set of boxes to add the hypothesized values for the proportions. Enter the null hypothesis values there; for example, we could ask whether the Bumpus data has equal representation of males and females, so we add 0.5 and 0.5 to the two boxes. Next, click one of the lines that say "Binomial test." Click "Done", and the results appear. 2. Using "Frequency" There are two ways that JMP can take frequency data. The first we have already seen: each row represents an individual, and each individual has a column with a categorical variable that describes which group that individual belongs to. Alternatively, in some cases it can be quicker to use a different format. If there are multiple individuals that are identical (for example, if we had a data set that recorded sex only, the rows would either be male or female only). In these cases, we can create a row that describes those individuals, and then add a column that records how many individuals are like that -- the frequency. For example, the sex of the sparrows could be in a data file that looked like this: 30 Biology 300 lab manual, Lab 5 In case like this, when we start to analyze the data, we need to tell the program which column has this frequency information. We can do this in either the "Distribution" or "Fit Y by X" menus, but adding the column with frequency data to the blank for "Frequency". 3. Goodness of fit tests A χ2 goodness of fit test can also be done from the "Distribution" window. Follow the instructions for a binomial test to choose "Test Probabilities" from the red triangle menu. After entering the expected proportions for each category, click on "probabilities not equal to hypothesized values (two-sided chi-square test)". (Note: for JMP, the hypothesized (or expected) values can be entered either as proportions or counts. In the results listing, the χ2 results will be shown in the row labeled "Pearson" (after the inventor of the χ2 goodness of fit test, Karl Pearson). For example, fitting the male/female data from the Bumpus data set to a 50:50 model, the P-value from the χ2 test is 0.0011. Note that when JMP says “Prob>Chisq” in the bottom right corner of this box, this is telling you the P-value of the test, or 0.0011 for Pearson’s χ2 test. You don’t need to look up the χ2 value in a statistical table; JMP has already found the Pvalue for you. 31 Biology 300 lab manual, Lab 5 A goodness of fit test can be done using the "Frequency" option described in point 2, as well. 4. Testing fit to a Poisson distribution JMP can be used to test the goodness of fit to specific probability distributions. For example, we may want to compare a set of data to the expectations of a Poisson distribution. In this case, the null hypothesis is that the data are drawn from a Poisson distribution. Open the file “cardiac events.csv”. This file records the number of heart attacks occurring per week in a hospital. The column titled “Number of events per week” is the data showing number of heart attacks in a given week. The column “Frequency - in-hospital cardiac events” shows the number of weeks that had that number of events. For example, there were ten weeks during this time period in which there were exactly five out-ofhospital cardiac events. There are a total of 261 weeks in the data set. To ask whether these data fit a Poisson distribution, choose “Distribution” from the “Analyze” menu. The variable we are wanting to look at is “Number of events per week”, so put that under “Y, columns”. The total number of data points that have that value of Y is listed in “Frequency - inhospital cardiac events”, so put that column into the “Frequency” box. The box should look this, and then click “OK”. In the window which opens in response, click on the red triangle next to “Number of events per week” and choose “Discrete Fit” and then “Poisson”: 32 Biology 300 lab manual, Lab 5 This will generate a Poisson distribution which has the same mean as the data and plot it over the histogram. It will also open a new section at the bottom of the window called “Fitted Poisson”. Click on the red arrow next to “Fitted Poisson” and choose “Goodness of Fit”. This will tell JMP to run a Goodness of Fit test to the Poisson distribution. In this case it tells us that “Prob > X2” is 0.7379, meaning that the P-value is 0.7379 for this test. 33 Biology 300 lab manual, Lab 5 Questions 1. Most hospitals now have signs posted everywhere banning cell phone use. These bans originate from studies on earlier versions of cell phones. In one such experiment, out of 510 tests with cell phones operating at near-maximum power, six disrupted a piece of medical equipment enough to hinder interpretation of data or cause equipment to malfunction. A more recent study found zero instances of disruption of medical equipment out of 300 tests. a. For the older data, calculate the estimated proportion of equipment disruption. What is the 95% confidence interval for this proportion calculated by JMP? b. For the data on the later cell phones, use JMP to calculate the estimate of the proportion and its 95% confidence interval. [Note: JMP, like most computer packages, uses the Wald method to calculate confidence intervals for proportions, which is not very accurate for proportions close to zero or one.] 2. It is difficult to tell what other people are thinking, and it may even be impossible to find out how they are thinking by asking them. A series of studies shows that we do not always know ourselves how our thought processes are carried out. A classic experiment by Nisbett and Wilson (1978) addressed this issue in a clever way. Participants were asked to decide which of four pairs of silk stockings were better, but the four stockings that they were shown side-by-side were in fact identical. Nearly all participants were able, however, to offer reasons that they had chosen one pair over the other. They were presented randomly with respect to which was in which position. However, of the 52 subjects who selected a pair of stockings, 6 chose the pair on the far left, 9 chose the pair in the left-middle, 16 chose the pair in the rightmiddle, and 21 chose the pair on the far right. None admitted later that the position had any role in their selection. Do a hypothesis test to ask, was the selection of the stockings independent of position? If so, describe the pattern seen. 3. Are cardiac arrests (or "heart attacks") equally likely to occur throughout the year? Or are some weeks more likely than others to produce heart attacks? One way to look at this issue is to ask whether heart attacks occur according to a model where they are independent of each other and equally likely at all times -the Poisson distribution. The data file "Cardiac events.csv" contains data from one hospital over five years. It records both heart attacks that occurred to individuals outside of the hospital and also heart attacks that occurred to patients already admitted to the hospital. 34 Biology 300 lab manual, Lab 5 Does the frequency distribution of out-of-hospital cardiac events follow a Poisson distribution? 4. Many people believe that the month that person is born in can predict significant attributes of that person in later life. Such astrological beliefs have little scientific support, but are there circumstances in which birth month can have a strong effect on later life? One prediction is that elite athletes will disproportionately be born in the months just after the age cutoff for separating levels for young players. The prediction is that those athletes that are oldest within an age group will do better by being relatively older, and therefore will gain more confidence and attract more coaching attention than the relatively younger players in their same groups. As a result, they may be more likely to dedicate themselves to the sport and do well later. In the case of soccer, the cutoff for different age groups is generally August. a. The birth months of soccer players in the Under-20's World Tournament are recorded in the data file "Soccer births.csv." Plot these data. Do you see a pattern? (Photo from VensPaperie on flickr.) b. The numbers of people born in Canada by month is recorded in the file “NHL births.csv” (don’t be confused by the fact that this file also has data on birth months of hockey players). Compare the distribution of birth months of the soccer players to what would be expected by chance, assuming that the birth data for Canada is a good approximation for the population from which soccer players are drawn. Do they differ significantly? Describe the pattern. 5. Return to the page from the activities section where you circled the letter "t" in the paragraph from Darwin's Origin of Species. (If you haven’t already done this, please stop reading here now and go back to do that first.) The point of this exercise is to collect data on whether our brains perceive words merely as a collection of letters or if sometimes our brains process words as entities. The logic of this test is that, if words are perceived as units, rather than as collections of letters, then we should be more likely to do so for common words. Hence we will look at the errors made in scanning this paragraph, and ask whether we are more (or less) likely to miss finding a "t" when it is part of a common word. 35 Biology 300 lab manual, Lab 5 Compare your results to the answer key that your TA will provide that marks all the instances of the letter "t". Note that the answer key marks all "t"s in red, but it also puts boxes around some words. The boxes are drawn around all instances of the letter "t" occurring in common words. "Common" is defined here as among the top-twenty words in terms of frequency of use in English; of these six contain one or more "t"s: the, to, it, that, with, and at. In this passage there are 94 "t"s, and 29 are in common words. Count how many mistakes you made finding "t"s in common words and in less common words. Use the data you collect yourself; also please report your results to the TA so that they can be analyzed with the rest of the class. a. If mistakes are equally likely for each common and less common word (as defined above), what fraction of mistakes would be predicted to be with common words, using this passage? b. Use the appropriate test to compare your results to the null expectation. 36 Biology 300 lab manual, Lab 6 6. Contingency analysis Goals Test for the independence of two categorical variables. Quick summary from text (Chpt 9 from Whitlock and Schluter) A contingency analysis allows a test of the independence of two or more categorical variables. The most common method of contingency analysis is based in the χ2 Goodness-of-Fit test. It makes the same assumptions as that test in terms of the minimum numbers of expected values per group. Fisher's Exact Test allows contingency analysis on two-by-two tables, where each variable has two possible categories. This test makes no assumptions about the minimum expected values. Learning the Tools 1. χ 2 contingency analysis A contingency analysis can be done from the "Fit Y by X" window. Choose one of the categorical variables for "X, Factor" (preferably the one that you view as the explanatory variable), and the other for "Y, Response." JMP will then plot a mosaic plot, as you have seen before, and below the mosaic plot are the results of a contingency analysis in the section marked “Tests”. The row marked "Pearson" gives the standard χ2 contingency analysis result. (The row marked "Likelihood Ratio" gives the results of a G-test. See section 9.5 in Whitlock and Schluter.) One thing that JMP does not do is very important. JMP does not check that the assumptions of the χ2 test are met. Specifically, it doesn’t check that the expected values are at least 5 for at least 80% of the cells. (Also it is necessary that all cells have an expected value greater than one.) If these assumptions are not met, ignore the χ2 and likelihood results, and use Fisher's exact test, given at the bottom of the window. To see the expected values, use the Fit Y by X window. Click on the red triangle next to “Contingency Table” and choose the option “Expected.” The expected values will appear at the bottom of each cell. 37 Biology 300 lab manual, Lab 6 2. Using "Frequency." As for the goodness of fit test, in JMP one can use a faster data format that summarizes the frequency of individuals that all share the same characteristics. For example, using the "Titanic" data, we could do a contingency analysis comparing the sex of passengers to whether they survived the wreck. In the way the data is already formatted, each individual has his or her own row. Alternatively, these data could be represented in the following data table: When using a data table like this, the column "Number in category" (or whatever it is that you name the column with that the frequency data) must be added to the "Frequency" box on the "Fit Y by X" screen. This approach will give exactly the same answers as the individual-based method. (Try it on these data to see.) 3. Calculating odds ratios Odds ratios (and other methods of describing the relationship between two categorical variables) can be calculated in JMP. After plotting the mosaic plot with "Fit Y by X," choose "Odds Ratio" from the red triangle menu at the top left of the window. The odds ratio and its 95% confidence interval will appear at the bottom of the window. 38 Biology 300 lab manual, Lab 6 Activities We'll do a straightforward exercise to see how χ2 behaves when the null hypothesis is true, and then later we'll see what χ2 looks like when the null hypothesis is false. To do so, let's start by using some dice to create a sample. The variables are boring here, but the point is to see how the process works. We'll have two variables: whether a die rolls a ONE, and the hand used to roll the dice. We know pretty much already that there should be no association between which hand is used to roll a die and the outcome. So if we test a null hypothesis that "hand" and "one" are independent, we ought to find that there is no association. In other words, the null hypothesis of independence is true. 1. Roll 30 dice (or one die 30 times) with each hand, and record the number of "one"s for each hand. Create a JMP file to record these data with two columns: Hand ("left" or "right") and One ("yes" or "no"). 2. Do a χ 2 contingency analysis on the results. Most samples ought to not reject the null hypothesis, but about one in twenty samples will reject the null hypothesis. In all of these cases the null hypothesis will be true, but by chance 5% of the samples will get a P-value less than 5%. 3. Repeat steps 1 and 2 many, many times. You can do this by hand, but we recommend using a java applet to simulate the data collection to speed it up. Go to http://onlinestatbook.com/stat_sim/contingency/index.html and, after it loads, click "Begin." This will open a window that will simulate this kind of data. Let's consider "left-handed rolls" as "Condition 1" and "right-handed rolls as "Condition 2." The column P(S) is where we enter the desired probability for each case. Enter 0.1666 (i.e. one-sixth) in the box for P(s) for both "conditions" in the top left. Enter 50 in both boxes for sample sizes there as well. The window should look something like this, with four boxes changed from their defaults: 39 Biology 300 lab manual, Lab 6 Now click the box called "Simulate 1." This will do the same kind of sample that you just did with the dice, with the results appearing in the top right corner. This applet will also calculate the χ2 for you. Now click "Simulate 5000" one or more times. This will make 5000 separate samples, and calculate a χ2 for each. The program will keep track of how many tests were statistically significant and how many were not (see the bottom right sector of the window). You'll probably find that about 5% of the results are "significant," meaning that the null hypothesis would have been rejected. This is as expected; the significance level of 5% mean that we expect 5% of tests of true null hypotheses to reject the null. 4. Change the true proportion of one of the P(s) in the simulation. Using the applet still, let's simulate a loaded die, where the dice in the rolled in the right hand are much more likely to roll a one. Let's change the probability in the first row to be 0.35, but leave the second row set at 0.1666. Keep the sample size the same. With these settings, the null hypothesis tested by the contingency analysis is now false: there is a different probability of success in the two different conditions. Before running the simulations, think for a moment about what you would expect the results to look like. Should more or fewer of the samples give high χ2 values and reject the null hypothesis? Will all samples reject the null hypothesis, given that the null hypothesis is false? Now run 5000 or so simulations. Compare what you see to what you expected. 40 Biology 300 lab manual, Lab 6 Questions 1. Child seats have been a great success in reducing injuries and fatalities to infants and toddlers under the age of two. This success has encouraged governments to require child seats for older children as well. The data file called "child seats-restraints only.csv" contains data about the consequences of car accidents when children ages two to six are involved in an accident severe enough that at least one other person is killed (Levitt 2005). This data file includes data on kids who were in car seats, who were in lap and shoulder belts, and who were in lap belts alone. The file excludes data on children who were wearing no restraint; the data is clear that wearing no restraint is substantially more dangerous that any of these other options. The column called "Severity" indicates how severe the injuries are to the child. The "Serious" category includes both death and incapacitating injuries. a. Using these data, is there a significant difference in the severity of the result between different methods of restraint? b. Describe the results qualitatively. What policy decisions should be made on the basis of these data? c. Calculate the odds ratio for Severity, comparing child seats to lap belts only. 2. Caribbean spiny lobsters normally prefer to live in shelters with other lobsters. However, this may be a bad idea if the other lobster is infected with a contagious disease. An experiment was done (Behringer et al. 2006) to test whether lobsters can detect disease in other lobsters. Healthy lobsters were given a choice between an empty shelter and a shelter containing another lobster. Half of the experiments were done when the other lobster was healthy, and in the other half the other lobster was infected with a lethal virus (but before it showed symptoms visible to humans). Of 16 lobsters given a choice between an empty shelter and one with a healthy other lobster, seven chose the empty shelter. However, of 16 given a choice between the empty shelter and sharing with a sick lobster, 11 chose the empty shelter. Is there statistical evidence that the lobsters are avoiding the sick lobsters? 3. Return to the data you collected on your class during the first week. Are dominant hand and dominant foot correlated? 41 Biology 300 lab manual, Lab 6 4. Human names are often of obscure origin, but many have fairly obvious sources. For example, "Johnson" means "son of John," "Smith" refers to an occupation, and "Whitlock" means "white-haired" (from "white locks"). In Lancashire, U.K., a fair number of people are named "Shufflebottom," a name whose origins remain obscure. Before children learn to walk, they move around in a variety of ways, with most infants preferring a particular mode of transportation. Some crawl on hands and knees, some belly crawl commando-style, and some shuffle around on their bottoms. A group of researchers decided to ask whether the name "Shufflebottom" might be associated with a propensity to bottom-shuffle. To test this, the compared the frequency of bottom-shufflers among infants with the last name "Shufflebottom" to the frequency for infants named "Walker." (By the way, this study, like all others in these labs, is real. See Fox et al. 2002.) They found that 9 out of 41 Walkers moved by bottom-shuffling, while 9 out of 43 Shufflebottoms did. Is there a significant difference between the groups? 5. Falls are extremely dangerous for the elderly; in fact many deaths are associated with such falls. Some preventative measures are possible, and it would be very useful to have ways to predict which people are at greatest risks for falls. One doctor noticed that some patients stopped walking when they started to talk, and she thought that the reason might be that it might be a challenge for these people to do more than two things at once. She hypothesized that this might be a cue for future risks, such as for falling, and this led to a study of 58 elderly patients in a nursing home. Of these 58 people, 12 stopped walking to talk, while the rest did not. Of the people who stopped walking to talk, 10 had a fall in the next six months. Of the other 46, 11 had a fall in that same time period. a. Do an appropriate hypothesis test of the relationship between "stops walking while talking" and falls. b. What is the odds ratio of this relationship? 42 Biology 300 lab manual, Lab 7 7. Normal distribution and inference about means Goals Visualize properties of the normal distribution. See the Central Limit Theorem in action. Calculate sampling properties of means. Quick summary from text (Chpts 10 + 11 from Whitlock and Schluter) The normal distribution is the "bellshaped curve." It approximately describes the distribution of many biological variables. The normal distribution has a single mode equivalent to its mean, and it is symmetric around the mean. The normal distribution has two parameters, the mean and the variance. If a distribution is approximately normal (bell-shaped), then about twothirds of the data will fall within one standard deviation above or below the mean. About 95% will all within two standard deviations above or below the mean. The Central Limit Theorem states that the sum or mean of a set of independent and equally distributed values will have a normal distribution, as long as enough values are added together. This means that the mean of large sample will be normally distributed, even if the distribution of the variable itself is not normal. The confidence interval for a mean assumes that the variable has a normal distribution in the population and that the sample is a random sample. The 95% confidence interval for the population mean is approximately 2 standard errors above and below the sample mean. The one-sample t-test is a hypothesis test that compares the mean of a population to a hypothesized constant. The one-sample t-test makes the same assumptions as the confidence interval. 43 Biology 300 lab manual, Lab 7 Activity 1: Give the TA the finger(s) Using "Student Data Sheet 7" at the end of this manual, please make the suggested measurements and record them on the sheet. Then, if you don't mind sharing these data with the class, give the sheet to your TA who will compile them for use in Question 2. Activity 2: Distribution of sample means We return to the applet we used in chapter 3, located at http://onlinestatbook.com/stat_sim/sampling_dist/index.html. We want to investigate three claims made in the text. Claim 1: The distribution of sample means is normal, if the variable itself has a normal distribution. First, hit "Animated" a few times, to remind yourself of what this applet does. (It makes a sample from the distribution shown in the top panel. The second panel shows a histogram of that sample, and the third panel shows the distribution of sample means from all the previous samples.) Next, hit the "10,000" button. This button makes 10,000 separate samples at one go, to save you from making the samples one by one. Look at the distribution of sample means. Does it seem to have a normal distribution? Click the checkbox by "Fit normal" off to the right, which will draw the curve for a normal distribution with the same mean and variance as this distribution of sample means. Claim 2: The standard deviation of the distribution of sample means is predicted by the standard deviation of the variable, divided by the square root of the sample size. The standard deviation of the population distribution in the top panel is given in the top left corner of the window, along with some other parameters. The sample size is set by the pull-down menu on the right of the sample mean distribution. (The default when it opens is set to N=5.) For N=5, have the applet calculate 10,000 sample means as you did in the previous exercise. If the sample size is 5 and the standard deviation is 5.0 (as in the default), what do you predict the standard deviation of the sample means to be? How does this match the simulated value? Change the sample size to N=25, and recalculate 10,000 samples. Calculate the predicted standard deviation of sample means, and compare it to what you observed. 44 Biology 300 lab manual, Lab 7 Claim 3: The distribution of sample means is approximately normal no matter what the distribution of the variable, as long as the sample size is large enough. (The Central Limit Theorem). Let's change the distribution of the variable in the population. At the top right of the window, change "Normal" to "Skewed." This will cause the program to sample from a very skewed distribution. Set N=2 for the sample size, and simulate 10,000 samples. Does the distribution of sample means look normal? Is it closer to normal than the distribution of individuals in the population? Now set N=25 and simulate 10,000 samples. How does the distribution of sample means look now? It should look much more like a normal distribution, because of the Central Limit Theorem. (Explain in your own words why this is so.) Finally, look at the standard deviations of the distribution of sample means for these last few cases, and compare them to the expectation from the standard deviation of individuals and the sample size. If you want to play around with this applet, it will let you draw in your own distribution at the top. Just hold down the mouse button over the top distribution, and it will let you paint in a new distribution to use. Try to make a distribution as non-normal as possible, and then draw samples from it. Learning the Tools 1. One-sample t-tests For an example, let's use the data in "titanic.csv" on the passengers of the Titanic. We'll ask whether the mean age of passengers was significantly different from 18 years old. The first step is to use the "Distribution" window to create a histogram and summary statistics. (You find "Distribution" under the "Analyze" menu bar.) Choose "age" as the variable, and hit OK. To do a one-sample t-test, click on the red triangle next to the variable name, "age." From the menu that appears, choose "Test Mean." A window will open that asks for the mean proposed by the null hypothesis. In our case, that value should be 18. Enter the value and click OK. 45 Biology 300 lab manual, Lab 7 This will open a new section in the results window, which will give the results of a one-sample t-test, with the value of the test statistic t, the degrees of freedom, and the P-value for one- and two-sided tests. 2. Checking for normality One of the best ways to know whether a population is approximately normal is to plot a histogram of the data. If the sample size is large and the data look even approximately normally distributed, then that will be close enough for most purposes. JMP will let you draw a normal distribution with the same mean and standard deviation as the data on top of the histogram, to make it easier to visualize. In the menu under the red triangle by the variable name, click "Continuous Fit" and choose "Normal" from that menu. To more formally look at normality, make a quantile plot. After you used "Continuous Fit," a new section called "Fitted Normal" will have appeared at the bottom of the window, with a new red triangle . Under that triangle, click "Diagnostic Plot." If a data set is approximately normal, then the points in the quantile plot will mainly fall along a straight line. 46 Biology 300 lab manual, Lab 7 These age data from the Titanic passengers are significantly different from a normal distribution. However, the shape of the distribution is not that different from a normal distribution, as you can see because the points mainly fall along the diagonal line in this quantile plot. Therefore, most methods that assume normality would work well in this case. 3. Transformations In some cases a distribution will be more normal if the data are transformed before analysis, for example by taking the log of each data point. To do so, make a new column in the data file, and in this column use Formula to create a transformation. For example, to do a logtransformation, in the Formula window choose "Log" under the Transcendental menu (under Functions), and then click on the variable you want to log to put it into the formula. For example, to log-transform age, here is what the formula will look like: After the formula is entered, hit "Apply" and "OK". You can then analyze the new variable just like any other variable in JMP. 47 Biology 300 lab manual, Lab 7 Questions 1. Insects like the fruit fly Drosophila melanogaster have immune systems, but they rely on innate immunity rather than acquired resistance to specific antigens. As such, the genetic variation in immune response is extremely important for understanding the effects of disease on fly populations. A large number of flies were measured for their ability to suppress growth of a gram-negative pathogenic bacteria. Each fly was inoculated with the disease, and then the amount of bacteria in each was measured later. The log-bacteria per fly is listed under the column “Bacterial load” in the file “antibacterial immunity.csv”. a. Use at least two methods to determine whether bacterial load is approximately normally distributed. b. On average, is the mean of the bacterial load score different from zero? Do the appropriate statistical test. c. Calculate a 95% confidence interval for the mean bacterial load score of these flies. d. Use the mean +/- 2 standard error approximation to calculate a 95% confidence interval for the mean score. Compare the result of this approximation to the more precise answer from part (b). 2. Earlier this lab you will have measured your fingers. We're going to use this data now and also in the next lab. We'll look at the ratio of the lengths of index finger over ring finger. This is called the 2D:4D ratio, where the 2D refers to second digit and 4D means fourth digit. It turns out—bizarrely—that this 2D:4D ratio is a measure of the amount of testosterone that a fetus is exposed to during gestation. Next week we'll test whether that corresponds to differences for males and females. For this week, we'll estimate the 2D:4D ratio, and ask whether its mean is significantly different from 1. If index fingers are on average equal to ring fingers in length, then the ratio would be one. First, use the finger data file your TA has compiled. Add two new columns, and in these columns use "Formula" to create the new variables "Right 2D:4D ratio" and "Left 2D:4D ratio." a. What is the 95% confidence interval for "Right 2D:4D ratio"? For the left ratio? 48 Biology 300 lab manual, Lab 7 b. For each of the two ratios, test whether the mean ratio differs from one. c. Compare the results from the confidence interval and the hypothesis test. Are they consistent? 3. Mosquitoes find their victims in part by odor, so it makes sense to wonder whether what we eat and drink influences our attractiveness to mosquitoes. A study in West Africa (Lefèvre et al. 2010), working with the mosquito species that carry malaria, wondered whether drinking the local beer influenced attractiveness to mosquitoes. They opened a container holding 50 mosquitoes next to each of 25 alcohol-free subjects and measured the proportion of mosquitoes that left the container and flew toward the subject (they called this proportion the “activation”). They repeated this procedure 15 minutes after each of the same subjects had had a liter of beer, and measured the “change in activation” (after minus before). The data in the file “BeerAndMosquitos.csv” are the values of mosquito activation (fraction of 50 mosquitos that left the container) recorded on the 25 subjects before and after drinking 1 liter of the local beer. a. Calculate a 95% confidence interval for the difference in activation after drinking beer. b. Does having beer change the average attractiveness of a person to mosquitoes? Do an appropriate hypothesis test. 4. The file "mammals.csv" contains information on the body size and brain size of various mammal species. a. Plot the distribution of brain size, and describe its shape. Does this look like it has a normal distribution? b. Use a histogram to examine whether brain size has a normal distribution. c. Transform the brain size data with a log-transformation. Plot the distribution of log brain size. Describe the new distribution, and examine it for normality. Assignment (this may change—see instructors for exact assignment) Using the data collected by all individuals in your class in response to the Activity in Lab 5, test whether there is evidence that people in general are likely to read common words as blocks rather than as a collection of letters (in other words, are "t"s in common words found differently than those in uncommon words). Write up the results in a journal article format, with an introduction, methods and materials, results, and discussion. Include both graphical and statistical representations of the data. The report should be approximately 3-4 double-spaced pages. The TA will give you a copy of the data for the whole class to work from. 49 Biology 300 lab manual, Lab 8 8. Comparing two groups Goals Compare the means of two groups Understand paired vs. two-sample designs Explore the effects of violation of assumptions Quick summary from text (Chpt. 12 from Whitlock and Schluter) The most common method to compare the means of two groups is the two-sample t-test. The two-sample t-test assumes that there is an independent random sample from each group, and that the variable is normally distributed in each population with the same variance. If each data point in one group is uniquely paired with a data point in the other group, then the data are said to be paired. A paired t-test examines the mean difference between the members of each pair. Paired t-tests assume that the differences are normally distributed and that the pairs are randomly sampled. Welch's t-test is similar to a two-sample t-test, but does not require the assumption of equal variances. Most hypothesis tests make assumptions about the distribution of the population that the data are taken from, for example, that the variable has a normal distribution. Such tests are called parametric tests. Many parametric tests, including the t-test, are robust to their assumptions This means that the test works well even when the assumptions are violated. For example, with large sample sizes, the 2-sample t-test works acceptably even if the two groups’ variances differ by a factor of 10. Data can sometimes be transformed to make new variables that better match the assumptions of the methods. For example, with the logtransformation, the natural logarithm of the value of a variable is used instead of the variable itself. 50 Biology 300 lab manual, Lab 8 Activities 1. Collect some data on your dexterity Using "Student Data Sheet 8", at the end of this manual, collect the data on the speed of each hand, and record them on the sheet. Then, if you don't mind sharing these data with the class, give the sheet to your TA who will compile them for use in Question 3. 2. Investigating robustness The t-test is fairly robust to its assumptions. This means that even when the assumptions are not correct, the t-test often performs quite well. Remember that the two-sample t-test assumes that the variables have a normal distribution in the populations, that the variance of those distributions is the same in the two populations, and that the two samples are random samples. Open a browser and load the applet by clicking "Begin" at http://onlinestatbook.com/stat_sim/robustness/index.html. This applet will simulate t-tests from random samples from two populations, and it lets you determine the true nature of those populations. Start with a scenario that matches the assumptions of the t-test. Use the default parameters of Mean = 0, standard deviation (sd) = 1, and no skew for both populations. (These values can be changed by the controls on the left hand side of the window.) You can leave the sample sizes at 5 (controls for the sample sizes are in the right side of the window.) Now click "Simulate" at the bottom. The computer will artificially create 2000 samples of 5 individuals each from both populations, and calculate the results of a two-sample t-test comparing the means. It tallies the numbers of significant and non-significant results at the bottom of the window. In this case, both distributions are normal and the variances are equal, just as assumed by the t-test. Therefore we expect it to work quite well in this case. (Take a moment to think about what "working well" means. If the null hypothesis is true, then the Type I error rate should match the stated Type I error rate, and if the null hypothesis is false it should be rejected as often as possible.) Now, look at the following cases, and describe the effects on the performance of the test. a. (Unequal standard deviation, equal sample size) Make the standard deviations of the two populations unequal, with the first equal to one and the second equal to 5, with everything else like the defaults. (In particular, keep the sample sizes equal.) 51 Biology 300 lab manual, Lab 8 b. (Unequal standard deviation, unequal sample size) Make the standard deviations unequal as in part a. (that is, 1 and 5), but make the sample size of the first population be 25, leaving the second sample with 5 individuals. c. (Skew, equal sample size) Return to the defaults, except make both populations have "Severe" skew. Play around with other parameter combinations. What can you conclude about the robustness of the two-sample t-test? Learning the Tools 1. Paired t-test To do a paired t-test in JMP, the data have to be stored in a particular way. Each pair needs to have a single row, with the value of one member in the pair in one column and the value for the other member in another column. For example, if the paired t-test was comparing a variable before and after a treatment, there would be one column for the variable before treatment and another column for after. There are two different ways to do a paired t-test in JMP. The first option is to create a new column and use "Formula" to calculate the difference between the two columns for the variable, for example after minus before. The difference column can then be analyzed with a one-sample t-test using the "Distribution" window." (This works because a paired t-test is just a one-sample t-test on the differences.) JMP will also do a paired t-test directly, using the "Matched Pairs" window, available under the "Analyze" menu bar. Choose the two variables that represent the two members of each pair (for example "before" and "after") for the "Y, paired response" blank, and hit OK. The resulting graph is rather confusing, but in essence it plots the difference in values against the mean of the two members of each pair. The results of a paired t-test are given at the bottom of this window. For example, the data file "NZNoggins.csv" contains information on head sizes of New Zealand army recruits, measured either with metal calipers or a cheaper cardboard method. Each row represents a different soldier, measured in each of the two ways. The results show that the cardboard method is biased, giving on average larger values than the metal calipers. 52 Biology 300 lab manual, Lab 8 2. Making a dot plot Making a dot plot in JMP takes a little work. Let’s use the titanic data set, to make a dot plot of the age of those who survived and those who died. With the titanic data set open, choose “Fit Y by X” from the “Analyze” menu. Choose “Survived” as the X variable and “Age” as the Y variable, and hit OK. In the resulting window with the graph, choose “Means and Std Dev” from the menu at the red triangle, which will add the means and confidence bars on the graphs. Next, choose “Display options” from the red triangle menu, and select “Points jittered”. This will spread the points out a bit so that you can see more about where the data are. The result is not the best dot plot you have ever seen, but it gives a sense of the data. 3. Two-sample t-test To do a two-sample t-test, first plot the numerical variable for both groups using “Distribution” under the “Analyze” menu. Choose your numerical variable as Y, and put your group variable in the box by “ By”. This will plot a histogram separately for both groups. You can analyze the data with "Fit Y by X" from the "Analyze" menu. For example, let's compare passengers on the Titanic that survived to those that died, to ask whether they differ in age. (We might expect that they do, because of the social norm to put "women and children first.") Choose "Survived" as the X variable, and "age" as the Y variable in the "Fit Y by X" screen. Next, from the red triangle menu at the top of the window, choose "Means/ANOVA/Pooled t." [Important note: Confusingly, don’t choose "t Test," which will do a Welch's t-test, as we discuss in the next section.] 53 Biology 300 lab manual, Lab 8 The results section that this will create contains a lot of information. Here are the highlights: 4. Welch's t-test (not assuming equal variances) Welch's t-test does not assume that the variance is the same in the two groups. To do a Welch's t-test in JMP, first open the "Fit Y by X" window, but then choose "t Test" under the red triangle. The results section that opens looks like the following, including the phrase "assuming unequal variances." Alternatively, under the red triangle menu, choose "Unequal variances." This does a test for the equality of variances and performs Welch's t-test automatically, as shown in the following result screen. 54 Biology 300 lab manual, Lab 8 Questions 1. A common belief is that shaving causes hair to grow back faster or coarser than it was before. Is this true? Lynfield and McWilliams (1970) did a small experiment to test for an effect of shaving. Five men shaved one leg weekly for several months while leaving the other leg unshaved. The researchers measured the change in leg hair width and the hair growth rate on each of the legs. These data are given in "leg shaving.csv." a. Find the 95% confidence interval for the difference between shaved and unshaved legs for both variables. b. Perform a suitable hypothesis test to ask whether shaving affects hair width. c. Perform a suitable hypothesis test to ask whether shaving affects hair growth rate. 2. In the last lab, we collected data on the lengths of your second and fourth fingers (2D and 4D -- your TA will tell you where to find the data.) As mentioned last time, the 2D:4D ratio is a measure of the amount of testosterone that a human is exposed to as a fetus during the period of finger development. As such, we might expect there to be a difference between males and females in the 2D:4D ratio. Use these data to calculate the 2D:4D ratio (index finger length divided by ring finger length) on the right hands. a. Test for a difference in the mean 2D:4D ratio between men and women. b. What is the 95% confidence interval for the difference in means? 3. Return to the data collected early in the lab on dexterity of each hand, which should be collated by the TA. Each of you has measured your dexterity with each hand. Do a hypothesis test to compare average difference in the dexterity of the dominant hand compared to the less dominant hand. 55 Biology 300 lab manual, Lab 8 4. London taxi drivers are required to learn 25,000 streets within a 6-mile radius of central London -- a massive compendium of spatial information referred to as The Knowledge. Spatial information is thought to be processed in a part of the brain known as the posterior hippocampus. (This part of the brain is larger in bird and mammal species that use a great deal of spatial information, such as species that store caches of food.) Maguire et al. (2000) wanted to know whether this great skill in spatial navigation translated into changes in brain morphology in London cabbies. They used MRI to measure the cross-sectional area of the posterior hippocampus in two groups of cab drivers: one group who had been driving for less than 15 years, and another group who had been driving for more than 15 years. The data are in the file "hippocampus.csv." a. Test for a difference in the mean posterior hippocampus size between newer and older drivers. b. What is the 95% confidence interval for the difference in means? 5. In 1898, Hermon Bumpus collected data on one of the first examples of natural selection directly observed in nature. Immediately following a bad winter storm, 136 English house sparrows were collected and brought indoors. Of these, 72 subsequently recovered, but 64 died. Bumpus made several measurements on all of the birds, and he was able to demonstrate strong natural selection on some of the traits as a result of this storm. Bumpus published all of his data, and they are given in the file "Bumpus.csv." Test whether the birds that survived differ from the dead birds for each of the following traits: a. Total length b. Sex c. Skull width d. Beak and Head length e. Weight f. Age 56 Biology 300 lab manual, Lab 9 9. Comparing more than two groups: ANOVA Goals Compare the means of multiple groups using Analysis of Variance. Make post-hoc comparisons with the Tukey-Kramer test. Quick summary from text (Chpt. 15 in Whitlock and Schluter) Analysis of variance (ANOVA) compares the means of multiple groups. A significant ANOVA result implies that at least one group has a different mean from one other group. Tukey-Kramer tests allow comparison of each pair of means. ANOVA assumes equal variance of each group, each group has a normal distribution of the variable, and each population is randomly sampled. The Kruskal-Wallis test is a nonparametric test that compares multiple groups, often used in place of ANOVA when ANOVA's assumptions aren't true. Learning the Tools 1. ANOVA To do a one-way ANOVA, we can use the "Fit Y by X" routine from the "Analyze" menu bar. The categorical or group variable should be entered as "X, factors" and the response variable should be entered as "Y, response." When the results window opens with a dot plot of the data, click on "Means/Anova" under the red triangle menu in the top left corner. For example, let's use the "15e1KneesWhoSayNight.csv" data set that correspond to example 15.1 in Whitlock and Schluter. "Treatment" is the group variable to put in the "X" box and "shift" is the numerical response variable to put in as "Y." Doing the ANOVA gives the following window of results. 57 Biology 300 lab manual, Lab 9 2. Tukey-Kramer tests JMP 7: Tukey-Kramer tests can also be done from the menu under the red triangle. Click on "Compare means" and then on "All Pairs, Tukey HSD." This will open a results section that looks something like this: 58 Biology 300 lab manual, Lab 9 3. Kruskal-Wallis test A Kruskall-Wallis test can be performed under an alternative name, the Wilcoxon test. In the menu under the red triangle at the top left of the window, click on "Nonparametric" and then choose "Wilcoxon Test." A results section at the bottom of the window will show the results. The Pvalue of the test is given under the heading of "Prob >ChiSq." Questions 1. The pollen of the corn (maize) plant is known to be a source of food to larval mosquitoes of the species Anopheles arabiensis, the main vector of malaria in Ethiopia. The production of maize has increased substantially in certain areas of Ethiopia recently, and over the same time malaria has entered in to new areas where it was previously rare. This raises the question, is the increase of maize cultivation partly responsible for the increase in malaria? One line of evidence is to look for a geographical association on a smaller spatial scale between maize production and malaria incidence. The data set "malaria vs maize.csv" contains information on several high-altitude sites in Ethiopia, with information about the level of cultivation of maize (low, medium or high) and the rate of malaria per 10,000 people. a. Plot the relationship between maize production and the incidence of malaria. b. Do these data seem to match the assumptions of ANOVA? If not, do any simple transformations solve the problem? c. Test for an association between maize yield and malaria incidence. 2. The European cuckoo does not look after its own eggs, but instead lays them in the nests of birds of other species. We have seen previously that cuckoos sometimes have evolved to lay eggs that are colored similarly to the host birds' eggs. Is the same true of size -- do cuckoos lay eggs of different sizes in nests of different hosts? The data file "cuckooeggs.csv" contains data on the lengths of cuckoo eggs laid in a variety of other species' nests. a. Test for a difference between the host species in the size of the cuckoo eggs. b. Using a Tukey-Kramer test, which pairs of host species are significantly different from each other? 59 Biology 300 lab manual, Lab 9 3. Animals that are infected with a pathogen often have disturbed circadian rhythms. (A circadian rhythm is a daily cycle in a behavior or physiological trait that persists in the absence of time cues.) Shirasu-Hiza et al. (2007) wanted to know whether it was possible that the circadian timing mechanism itself could have an effect on disease. They used three groups of fruit flies: one "normal", one with a mutation in the timing gene tim01, and one group that had the tim01 mutant in a heterozygous state. They exposed these flies to a dangerous bacteria, Streptococcus pneumoniae, and measured how long the flies lived afterwards, in days. The date file "circadian mutant health.csv" shows some of their data. a. Plot a histogram of each of the three groups. (Hint: you can use the "By" function in the Distribution window to specify the group.) Do these data match the assumptions of an ANOVA? b. Do an appropriate hypothesis test to ask whether lifespan differs between the three groups of flies. 60 Biology 300 lab manual, Lab 10 10. Regression and correlation Goals Use the computer to calculate regression lines and correlation coefficients, test null hypotheses about slopes, and deal with non-linear data. Quick summary (Chpts 16 + 17 from Whitlock and Schluter) Linear regression uses a line to predict a response numerical variable (Y) from a numerical explanatory variable (X). The regression line will take the form Y = a + b X. a is the intercept (where the line crosses the axis at X = 0), and b is the slope of this regression line. Linear regression assumes that the true relationship between X and Y follows a line, that the data are taken from a random sample, and that for every value of X the corresponding values of Y are normally distributed with the same variance for all X. These assumptions can be examined with a residual plot, where the residual (deviation between the Y value and its predicted value) is plotted against X. Hypothesis tests about regression slopes can be made using either a t test or an ANOVA-like approach. Confidence bands show the limits of confidence intervals for the mean Y for each value of X. Prediction intervals show confidence intervals for the individual values of Y for each X. R2 measures the fraction of variance in Y that is predicted by X. Non-linear relationships between X and Y can often be examined by use of transformations. The correlation coefficient (r) measures the strength of association between two numerical variables, X and Y. A positive r means that larger values of X are associated with, on average, larger values of Y. A negative r means that large X values and small Y values tend to appear together. Hypothesis tests and confidence intervals for the correlation coefficient assume that X and Y have a linear relationship and that the sample used 61 Biology 300 lab manual, Lab 10 is a random sample. These methods also assume that for any given value of X, Y is normally distributed with equal variance. Also, for any value of Y, X is assumed to be normally distributed with equal variance. Learning the Tools 1. Linear regression We'll illustrate the steps of using the program to do regression with Example 17.1, predicting the age of a lion from the amount of black on its nose pad. In the data file, the age of lion is called age, given in years, and the proportion of black on its nose pad is called proportion.black. Load the data from the file "17e1LionAges.csv". Make a scatter plot of the relationship between age and proportion.black, using "Fit Y by X" from the Analyze menu bar. Have age be the response variable and proportion.black be the explanatory variable. To calculate the regression line and test the null hypothesis that the slope is zero, click on the small red triangle at the top left of the scatter plot window. From the menu that opens, choose "Fit Line." This will increase the size of the window, with new results added below the scatter plot. These results include the equation for the best-fitting line, the R2 value for the regression, and hypothesis tests of the null hypothesis that the slope of the line is zero. The hypothesis test is done both by the ANOVA approach and by using the t-test. 62 Biology 300 lab manual, Lab 10 2. Residual plots Next, examine the residuals plot for deviations from linearity and other assumptions. Click on the red triangle next to "Linear Fit," right under the graph. From the menu that appears, choose "Plot Residuals." A residual plot will appear at the bottom of the window. Look for evidence of unequal variance around the line or patterns that may indicate a non-linear relationship. For the lion data, there is no strong reason to doubt the assumptions of the analysis are OK. 63 Biology 300 lab manual, Lab 10 3. Transform data as required. This example does not require transformation. If data does need a transformation, you can easily do it using JMP. First, create a new column in the spreadsheet, and then use the "Formula" function to calculate transformed values in the new column. See chapter 7 for discussion of transformations in this way. 4. Calculate confidence bands or prediction intervals. Confidence bands for the mean predicted Y can be printed on the graph by choosing "Confid Curves Fit" from the same menu under the red triangle at "Linear Fit." Prediction intervals (showing the confidence intervals for predictions of individual values of Y) can fit by clicking "Confid Curves Indiv" from the same menu. The curves closest to the lines will be the confidence bands for the mean Y; those further from the line will indicate the individual prediction intervals. 64 Biology 300 lab manual, Lab 10 5. Calculating correlation coefficients The Pearson's correlation coefficient between two numerical variables can be calculated using the "Multivariate Methods Multivariate" menu choice under the "Analyze" menu. Choose the variables that you want to correlate by putting them in the "Y, columns" list, and hit OK. To make statistical tests about correlation coefficients, click on the red triangle by the "Multivariate" title, and choose "Pairwise Correlations." A results section will appear with the correlation between each pair of variables and the P-value of a test of the null hypothesis that they are not correlated. In this display, “Correlation” is the correlation coefficient r, and “Signif Prob" is the P-value for the test that this correlation coefficient is zero. Activities (Optional) Use the applet at http://onlinestatbook.com/stat_sim/transformations/index.html to explore the effects of transformations on the various data sets provided there. Click the transformation buttons on the side and bottom of each graph, and select alternative data sets from the menu button at the top. Questions 1. The ends of chromosomes are called telomeres. These telomeres are shortened a bit during each cell cycle as DNA is replicated; one of their purposes is to protect more valuable DNA in the chromosome from degradation during replication. As individuals age, their telomeres shorten, and there is evidence that shortened telomeres may play a role in aging. Telomeres can be lengthened in 65 Biology 300 lab manual, Lab 10 germ cells and stem cells by an enzyme called telomerase, but this is not expressed in most healthy somatic cells. (Cancer cells, on the other hand, usually express telomerase.) Given that the length of telomeres is biologically important, it becomes interesting to know whether telomere length is inherited. A set of data was collected by Nordfjäll et al. (2005) on the telomere length of fathers and their children; these data are in the file "telomere inheritance.csv." a. Plot a scatter plot showing the relationship between father and offspring telomere length. b. Do the data require any transformation before analysis? c. Create an equation that predicts the offspring telomere length from its father's. Is there evidence that the father's telomere length predicts his offspring's value? d. How many offspring values lie outside the prediction intervals for this regression line? Is this more than you would expect by chance? Explain. 2. Opfer and Segler (2007) asked several school children to mark on a number line where a given number would fall. The child was given a number line with two ends marked at 0 and 1000, and then asked to make an X on that line where a number, for example 150, should be placed. They asked the children to place several different numbers on the number lines, each on a fresh new piece of paper. Two separate groups of kids were asked to do this task: one group of secondgraders and another group of fourth-graders. The researchers then measured each mark to see the value on a linear scale of its placement. The results, averaged over all 93 kids for each group, are given in the file "numberline.csv". a. Plot the fourth graders' guesses against the true value. Is this relationship linear? If not, find a transformation of X or Y that describes the data well. b. Plot the second-graders' guesses against the true value. Is this relationship linear? If not, find a transformation of X or Y that describes the data well. Examine the residual plots for both the linear and nonlinear fits. 66 Biology 300 lab manual, Lab 10 c. Assume that the difference between the shapes of these curves is real. What would you conclude about the difference in the way 2nd graders and 4th graders perceive numbers? 3. Larger animals tend to have larger brains. But is the increase in brain size proportional to the increase in body size? A set of data on mammal body and brain size was collated by Allison and Cicchetti (1976), and theses data are to be found in the data set "mammals.csv." The file contains columns giving the species name, the body mass (in kg) and brain size (in g) of 62 different mammal species. a. Plot brain size against body size. Is the relationship linear? b. Find a transformation (for either or both variables) that makes the relationship between these two variables linear. c. Is there statistical evidence that brain size is correlated with body size? d. What line best predicts (transformed) brain size from (transformed) body size. e. If a mammal species had a log- body size that was three units greater, how much would log-brain size be expected to change? f. Make a residual plot. Which species has the highest brain size for their body size? Which species has the smallest brain relative to that predicted by its body size? g. Are humans significantly different from the brain size predicted by our body size? h. Are chimpanzees significantly different from the brain size predicted by their body size? 67 Biology 300 lab manual References and Photo Credits Chapter 1 countries data from : http://www.worldbank.org/ Muchhala, N. 2006. Nectar bat stows huge tongue in its rib cage. Nature 444:701-702. Chapter 3 McCusker, R. R., B. A. Goldberger, and E. J. Cone. 2003. Caffeine content of specialty coffees. Journal of Analytical Toxicology 27:520-522. Chapter 5 soccer birth data: Barnsley, R. H., A. H. Thompson, and P. Legault. 1992. Family planning: football style. The relative age effect in football. Int. Rev. for Soc. of Sport 27:77-86. NHL birth data: http://www.cd-b.com/nhl2-1.htm Birth months: Statistics Canada Cardiac arrest distribution: Skogvoll, E., and B. H. Lindqvist. 1999. Modeling the occurrence of cardiac arrest as a Poisson process. Ann. Emerg. Med. 33:409-17. Position effects: Nisbett, R. E., and T. D. Wilson. 1977. Telling more than we can know: verbal reports on mental processes. Psychological Review 84:231-259. Nisbett, R. E., and T. D. Wilson. 1978. The accuracy of verbal reports about the effects of stimuli on evaluations and behavior. Social Psychology 41: 118-131. Chapter 6 Levitt, S. D. 2005. Evidence that seat belts are as effective as child safety seats in preventing death for children aged two and up. Review of Economics and Statistics 90: 158-163. Behringer, D. C., M. J. Butler, and J. D. Shields. 2006. Avoidance of disease by social lobsters. Nature 441:421. Caribbean spiny lobster photo: http://www.flickr.com/photos/sniffette/1677217581/ Mohammed, M. A., J. Mant, L. Bentham, A. Stevens, and S. Hussain. 2006. Process of care and mortality of stroke patients with and without a do not resuscitate order in the West Midlands, UK. International Journal for Quality in Health Care 18:102–106. Fox, A. T., R. D. Palmer, and P. Davies. 2002. Do "Shufflebottoms" bottom shuffle? Arch. Dis. Child. 87:552-554. Lundin-Olsson, L., L. Nyberg, and Y. Gustafson. 1997. 'Stops walking when talking' as a predictor of falls in elderly people. Lancet 349:617. Chapter 7 Lazzaro, B. P., B. K. Sceurman, and A. G. Clark. 2004. Genetic basis of natural variation in D. Melanogaster antibacterial immunity. Science 303:1873-1876. Chapter 8 Lynfield, Y. L., and P. MacWilliams 1970. Shaving and hair growth. Journal of Investigative Dermatology 55:170-172. Maguire, E. A., et al. 2000. Navigation-related structural change in the hippocampi of taxi drivers. PNAS 97: 4398 – 4403. 68 Biology 300 lab manual Bumpus, Hermon C. 1898. Eleventh lecture. The elimination of the unfit as illustrated by the introduced sparrow, Passer domesticus. (A fourth contribution to the study of variation.) Biol. Lectures: Woods Hole Marine Biological Laboratory, 209-225. Chapter 9 Latter, O.H. 1902. The eggs of Cuculus canorus. An Inquiry into the dimensions of the cuckoo's egg and the relation of the variations to the size of the eggs of the foster-parent, with notes on coloration, &c. Biometrika i, 164. Kebede, A., J. C. Mccann, A. E. Kiszewski, and Y. Ye-Ebiyoam. 2005. New evidence of the effects of agro-ecologic change on malaria transmission. J. Trop. Med. Hyg. 73:676–680. Shirasu-Hiza, M., M. S. Dionne, L. N. Pham, J. S. Ayres, and D. S. Schneider. 2007. Interactions between circadian rhythm and immunity in Drosophila melanogaster. Current Biology 17: R353-R355. Chapter 10 Allison, T., and D. Cicchetti. 1976. Sleep in mammals: Ecological and constitutional correlates. Science 194:732-734. Nordfjäll, K., Å. Larefalk, P. Lindgren, D. Holmberg, and G. Roos. 2005. Telomere length and heredity: indications of paternal inheritance. PNAS 102:16374-16378. Opfer, J., and R. Siegler. 2007. Representational change and children's numerical estimation. Cognitive Psychology 55:169-195. brain/body size data available at http://lib.stat.cmu.edu/datasets/sleep. 69 Biology 300 lab manual Stats lab student data sheet 1 Record the following measurements on yourself. These will be shared (without your name attached) with the rest of the class, so if you feel any discomfort with such sharing feel free to omit any or all of the measurements. Height (cm) Circumference Measure around the of head (cm) head at eye-brow level. Sex Number of siblings How many siblings (not counting yourself) are in your family? Handedness Which hand do you normally write with? Dominant foot Which foot do you kick a ball with? Dominant eye Make a triangle with your index fingers and thumbs, and hold it out at arms' length. With both eyes open, focus on a specific spot in the distance, and center that spot in the triangle of your fingers. Now hold your arms still and shut one eye after the other. The spot will be closer to the center of the triangle when viewed with your dominant eye. 70 Biology 300 lab manual Stats lab student data sheet 7 Record the following measurements on yourself. These will be shared (without your name attached) with the rest of the class, so if you feel any discomfort with such sharing feel free to omit any or all of the measurements. Sex Handedness Which hand do you normally write with? Right index finger length Measured in mm from the basal crease of the finger to the tip -- see 2D in the figure.) This is called 2D, because it is on the second digit, counting the thumb. Right ring (4th) finger length Measured in mm from the basal crease of the finger to the tip-- see 4D in the figure. Left index finger length Same as above, but on left hand. Left ring (4th) finger length Same as above, but on left hand. 71 Biology 300 lab manual Stats lab student data sheet 8 Record the following measurements on yourself. These will be shared (without your name attached) with the rest of the class, so if you feel any discomfort with such sharing feel free to omit any or all of the measurements. Handedness Hand dexterity Right hand Hand dexterity Left hand Which hand do you normally write with? Take the test at http://faculty.washington.edu/chudler/java/rldot.html with each hand. Flip a coin to decide which hand to do first. 72 Biology 300 lab manual Tips for using JMP JMP is a very useful and mostly intuitive program. Here are a few tips to make it easier to use. Use the “Analyze” menu A great deal of what you want JMP to do for you starts in the “Analyze” tab of the main menu bar. Under this “Distribution” will help you visualize and describe individual variables, “Fit Y by X” will do most basic statistical analysis on two variables, and “Multivariate methods” will let you do correlations (and a lot of other things). Explore for the red triangles The analysis windows in JMP often include small red triangles like this . When you click on these a drop-down menu with many appropriate choices will appear. These choices let you do further analyses, change the features of graphs and tables, set key parameters, etc. Working with columns A column in JMP represents a variable, or the results of a formula that use variables from other columns. Column names and properties can be changed by double-clicking on the column header. You can change whether JMP thinks of a variable as continuous (i.e., numerical) or nominal (i.e. categorical) in this way. The “By” function Many analyses allow you to look at the data separately “by” a grouping variable. For example, in the “Distribution” window, if you use a variable “Sex” in the “By” box, then whatever analysis you ask for will be done separately for all the males and also all the females. Finding individuals The graphs in JMP are cleverly linked to the data file. If you click on a bar in a histogram, all the individuals that are part of that bar will be selected in the data file. If you click on a point in a scatterplot, that individual will be selected. Any selected individuals will then be marked in any graph associated with that data. Getting help JMP has a help button on the right of the main menu bar. It doesn’t always work smoothly. You may want to Google it with the word JMP added in the search. 73 Biology 300 lab manual Copying a graph (or anything else) from JMP If you want to use a graph for JMP in another file, it helps to know how to copy and paste from JMP's output. Use the “fat-plus” tool from the top of the analysis window. After you have clicked on the fat-plus, select the part of the window that you want to copy. Clicking on the section title usually works well. Then “Copy” from the “File” menu and paste it wherever you like. Finding P-values JMP has an idiosyncratic way of giving P-values. Instead of saying something like P < 0.05, or P = 0.017, JMP will have something that says, for example, Prob > ChiSq = 0.017, or Prob > t = 0.002, or something similar. In both cases, this is JMP's way of giving the P-value, P = 0.017 in the first case and P = 0.002 in the second case. Please note that JMP is calculating the P-value directly for you, so there is no need to take the test statistic to a stats table to get the P-value. Of course, doing this can be a great check on your understanding of both JMP’s output and how to use the stats tables. 74 Biology 300 lab manual Writing a Lab Report (compiled by BIOL 300 TAs) Format Include a brief descriptive title. 12 point font; Double spaced; 2cm margins; use a legible font. Figures need to communicate effectively (don’t make them too small). Don’t go over the page limit (which includes figures!) – points are deducted for pages over length. Write in paragraph form. Label each section with a heading (i.e. Introduction, Methods, etc.). Tone Avoid use of slang or casual abbreviations. Read other journal articles to get an idea of tone used. Note: section lengths given below for Introduction, Methods, Results and Discussion are suggestions, and not requirements. Introduction Introduce the main topics and explain why they are important and why we care about them. Define any important concepts/terms that will be referred to in the paper. Clearly state the question being addressed by your experiment. Give a brief overview of what you did to address the question. Mention the biological relevance or usefulness of the study. Start with broad conceptual information and narrow focus to the point of the study by the end of the intro. In this course, your intro could be ~1/2 – 1 page. Methods Summarize, in paragraph form, what you did. Choose an appropriate level of detail so that another scientist could repeat your study, but not so much detail that the reader is overwhelmed or disinterested (i.e., we don’t need to know who in the group did what, the column titles in JMP, etc.) 75 Biology 300 lab manual The methods should be in paragraph form, like the other sections. Typically in this course the methods might be ~1/2 a page. Results Report in paragraph form all findings that are relevant to your research question. Do not interpret your findings in this section; just state what you found. If necessary include figures or tables with complete captions. Every figure and table you include must be referred to in the text. Figures must include labeled axes (do this in JMP, or by hand) including units and scale. Figures should include legends when necessary. It is standard that figure captions are placed below the figure, whereas table captions are placed above the table. Captions should be detailed enough to stand alone. For example: Figure 1. The difference in circumference between cowrie shells measured by groups A&B. Mean = 2mm (+/- SE .3 mm) Text should come before the figures and in the text you should refer to your figures in numerical order. You should also present your figures in numerical order. When you report a mean, you must also report some measure of the uncertainty in the mean or variance in the population. In this course, text could be ~1/2 page, and altogether the figures ~ 3/4 to 1 full page. Discussion Discuss the implications of your Results. Explain why you think you found what you found. Relate your findings back to your initial question (from the Introduction). Did you find what you expected? If not, why not? Briefly summarize any benefits or problems with the design. Mention any relevant improvements. Finish with some general conclusions and potentially some ideas for more interesting follow up experiments In this course, the discussion might take ~1-1.5 pages. 76 Biology 300 lab manual General Avoid referencing the TA or saying things like “in the Bio 300 lab, etc.” There’s no need to talk about the details of how the data were stored (e.g., “we created columns in JMP labeled A1, A2, etc.”, unless this will make the methods easier to understand in some way. In the text, do not describe in detail the figures that are included. Just reference them and report what they are showing. For example, do not say things like “the histogram of X has the highest bar at a frequency of 0.4”... this can be easily seen by someone who looks at the figure. Every figure and table needs to be referenced in the main text. Avoid repeating the same word multiple times in a sentence. Avoid including arbitrary information (e.g., “We labeled group members A1, A2, B1, B2), unless this somehow makes things clearer later on. Have someone proofread to check for grammar, tense, spelling, paragraph structure and writing flow. 77