Restriction Analysis
advertisement

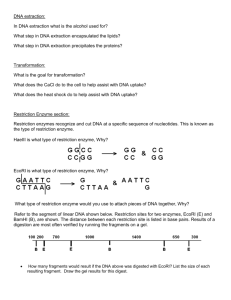
Restriction Analysis Eva Corinna Feil, Eva-Maria Neher, XLAB, Göttingen Restriction Enzymes estriction enzymes are very important enzymes that are vital to our manipulation of DNA. Restriction enzymes are proteins produced by bacteria to prevent or restrict invasion by foreign DNA. They act as DNA scissors, cutting foreign DNA into pieces so it cannot function. Restriction enzymes recognize and cut at specific sequences of nucleotides in the DNA molecule, called restriction sites. Each different restriction enzyme (and there are hundreds, made by many different bacteria) has its own restriction site. R A typical restriction site is 4-6 base pairs (bp) long. It is a palindrome; that is, it reads the same backwards and forwards. Along a DNA molecule, a specific combination of 4 bp occurs randomly once every few hundred bases. A specific sequence of 6 bp occurs randomly once every few thousand bases. Some DNA molecules have many sites for a particular restriction enzyme; others have none. When restriction enzymes cut DNA into pieces, the sizes of the DNA fragments correspond to the distances (in base pairs) between restriction sites. Restriction enzymes are named for the bacteria that produce them. SspI comes from the bacteria Sphaerotilus species (the Roman number I in the enzyme’s name shows that this was the first restriction enzyme found in this organism.) Pstl was the first restriction enzyme isolated from Providencia stuartii. And Hpal was the first restriction enzyme isolated from Haemophilus parainfluenzae. Restriction Maps and Logic Puzzles A new DNA molecule is like uncharted territory to a scientist. Like any explorer, the first thing a scientist does with new territory is to map its important features, such as size and the relative locations of certain parts. Scientists have a unique set of tools to map new DNA. Maps of DNA are called restriction maps, named for the enzymes used as mapping tools. Restriction enzymes recognize and cut DNA at specific sequences of nucleotides (called restriction sites), and the sizes of the DNA fragments correspond to the distances (in base pairs) between restriction sites. Every time the same piece of DNA is cut with a given enzyme, the same fragments are produced. Working out the locations of the different restriction sites is a problem in logic. Doing a restriction enzyme digest with a single enzyme, for example, only tells you how many sites are present for that enzyme. Doing restriction digests with more than one enzyme at a time can give clues as to where those restriction sites are in relation to each other. In the logic puzzle below, you have a series of step-by-step questions to help you make a restriction map of unknown DNA. First, you examine a mock gel to compare the DNA 53 fragments from restriction digests of the unknown DNA to DNA fragments of known sizes. Figure 1 shows lambda DNA digested with the restriction enzyme Pstl to produce DNA fragments of known sizes. Use the sizes of the known DNA fragments to estimate the sizes of the unknown DNA fragments. Then, add up the sizes of the individual fragments from each restriction digest to determine how large the original uncut piece of DNA was. Next, determine the Figure 1 number of times each restriction enzyme cut the DNA, and the distance between the cut sites. Finally, determine where the cut sites are relative to one another. Four different restriction enzyme digests have been done on our unknown DNA (Fig. 1). Three restriction enzymes have been used-Hpal, PstI, and Sspl. Samples of our DNA have been incubated with either Hpal alone; Hpal and PstI; Hpal and Sspl; or Hpal, PstI, and Sspl. The single restriction enzyme digest helps you deduce the number of cut sites present; the double and triple digests allow the cut sites to be mapped relative to each other. Questions 1. Estimate the sizes of the DNA fragments in base pairs (bp) by comparing them to the labeled lambda/Psd -size markers. These sizes do not have to be exact. Sizing of the smaller fragments is more accurate than sizing of the larger fragments. 2. Determine the total size of the digested DNA by adding up the sizes of the fragments from each digest. Take an average size from the 4 digests. Remember, the same DNA was digested in each sample, so the fragment sizes from the different digests should always add to the same total. 3. There are 2 Hpal sites present. Based on the number of fragments obtained from the Hpal digest, is this DNA linear or circular? Draw the DNA with the Hpal sites present. 54 4. How many PstI sites are present? 5. Where is the PstI site? Draw the position of the PstI site on the plasmid, relative to the Hpal sites. Remember, a plasmid is just a small, circular piece of DNA. 6. How many SspI sites are present? 7. Where is the SspI site? Draw the position of the SspI site on the plasmid, relative to the Hpal sites. It might be best if this is done in a separate sketch from the PstI site sketch, because we have not yet determined where the SspI and PstI sites are relative to each other. 8. Will the 600-bp Hpal fragment remain unchanged after digestion with either PstI or Sspl? (Check Fig. 1). 9. Which fragments are unchanged from the Hpal/Pstl digest to the Hpal/Pstl/SspI digest? Which fragments disappeared? Why did those fragments disappear? 10. Which fragments are unchanged from the Hpal/SspI digest to the Hpal/Pstl/SspI digest? Which fragments disappeared? Why did those fragments disappear? 11. Is there a fragment that appears only in the Hpal/Pstl/SspI digest? What does this mean? 12. Draw the full plasmid map, with all restriction enzyme recognition sites present in their relative locations. 55 Answers 1. Hpal 600 Hpal/PstI 600 1200 Hpal/SspI Hpal/Pstl/Sspl 300 600 300 600 1200 1800 2100 3000 3300 2. 3,900 base pairs (3.9 Kb) 3. This is circular DNA (plasmid). Restriction mapping can certainly be done on linear DNA; however, it is slightly more complex and calls for a different set of restriction enzyme digests. 4. Since there are 2 fragments after digestion with Hpal and 3 fragments after digestion with Hpal and PstI, there is one Pstl site. 5. The Pstl site is within the 3,300 bp Hpal fragment. 6. Since there are 2 fragments after digestion with Hpal and 3 fragments after digestion with Hpal and Sspl, there is 1 SspI site. 56 7. The SspI site is in the 3,300-bp Hpal fragment. 8. Yes. No SspI or Pstl sites are present within this fragment. 9. 600 and 1,200 bp. The 2,100-bp fragment disappeared because it contains a SspI site. 10. 300 and 600 bp. The 3,000-bp fragment disappeared so it must contain a Pstl site. 11. Yes. That there is a fragment with a SspI site on one end and a Pstl site on the other end. 12. Full-plasmid map: You may draw either of the 2 options; they are simply mirror images of one another. 57