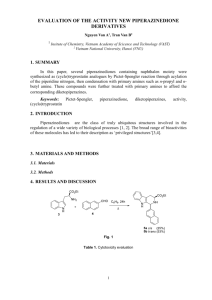
Hybrid Models for Combined Quantum Mechanical and Molecular
advertisement

10580 J. Phys. Chem. 1996, 100, 10580-10594 Hybrid Models for Combined Quantum Mechanical and Molecular Mechanical Approaches Dirk Bakowies† and Walter Thiel* Organisch-Chemisches Institut, UniVersität Zürich, Winterthurerstrasse 190, CH-8057 Zürich, Switzerland ReceiVed: December 8, 1995; In Final Form: March 13, 1996X A hierarchy of three models for combined quantum mechanical (QM) and molecular mechanical (MM) approaches is presented. They simplify the QM description of large molecules by reducing it to the electronically important fragment which interacts with the molecular mechanically treated remainder of the molecule. In the simplest model A, the QM fragments are only mechanically embedded in their MM environment. The more refined models B and C include a quantum mechanical treatment of electrostatic interactions between the fragments and a semiclassical description of polarization. The implementation of models A-C for MNDO type wavefunctions and the MM3 force field is outlined. Selected applications in organic chemistry are discussed, addressing the ability of the proposed models to reproduce substituent effects (MM) on chemical structure and reactivity (QM). These applications include protonations, deprotonations, hydride transfer reactions, nucleophilic additions, and nucleophilic ring cleavage reactions. 1. Introduction Quantum theory forms the physical basis for a large variety of computational approaches to chemical structure and reactivity. Due to the rapid development of computer hardware and software, small to medium-sized molecules can be studied by accurate ab initio methods.1 Density functional theory2 and semiempirical methods3 extend the range of possible applications: For example, semiempirical calculations are routine for systems up to about 100 atoms, and remain feasible for molecules up to about 1000 atoms.4,5 Nevertheless, there are many problems of chemical interest which cannot be handled by quantum mechanical treatments now or in the near future. These include studies of chemical reactivity in the presence of solvents and biochemical applications such as investigations of the catalytic effect of proteins in enzymatic reactions. The explicit description of the solvent or the protein leads to very large systems, and the use of quantum mechanical potentials in Molecular Dynamics (MD) or Monte Carlo (MC) simulations of such systems is clearly beyond the scope of current computational ressources. Empirical force fields6 have been employed successfully in many MD and MC simulations, but they are not suitable for problems of chemical reactivity since they do not allow an adequate description of the formation or cleavage of chemical bonds. This dilemma motivates the development of hybrid quantum mechanical (QM) and molecular mechanical (MM) models which combine the merits of both the quantum and the classical approach. The first is more generally applicable and allows the calculation of ground and excited state properties of electronic properties such as the ionization potential or the electron affinity and of chemical reactions. QM approaches require parametrization not at all (ab initio) or only to a limited extent (semiempirical), whereas the success of a classical force field strongly depends on the careful calibration of a large number of parameters against experimental reference data. This ensures the reliability and accuracy of a given MM approach, but also restricts its application to the classes of molecules it has been designed for. Many force fields outperform simple QM models in accuracy because specially tailored potential † Current address: Department of Pharmaceutical Chemistry, University of California, San Francisco, CA 94143-0446. X Abstract published in AdVance ACS Abstracts, May 15, 1996. S0022-3654(95)03651-3 CCC: $12.00 functions and flexible parametrizations allow a good reproduction of experimental data.6 A further advantage of MM force fields is their computational efficiency which makes them particularly attractive for MD or MC simulations. They are much faster than QM approaches and show a more modest scaling of the cpu requirements with the size of the system. The development of hybrid QM/MM approaches is guided by the general idea that large chemical systems may be partitioned into an electronically important region which requires a quantum mechanical treatment and a remainder which only acts in a perturbative fashion and thus admits a classical description. The perturbation may be mainly mechanical, e.g. if the classical region forces the quantum region into a particular geometry, but it may also include electronic effects such as electrostatics and polarization. According to this concept a chemical reaction is treated as a transformation involving only the reactive center (QM region) which is influenced by its environment (MM region, e.g. the solvent or a protein). Obviously this concept is closely related to common chemical argumentation. Several hybrid QM/MM models have been developed in the last two decades, combining semiempirical,7-12 density functional,13 valence bond,14,15 or ab initio Hartree Fock16 methodology with frequently used force fields. Most of the reported applications are devoted to solvation phenomena,17-31 while some deal with biochemical problems.32-36 Aiming for applications in organic chemistry (particularly organic reactions), we have coupled semiempirical quantum mechanical MNDO type methods37-39 with the molecular mechanical force field MM3.40,41 Both approaches are well established in organic chemistry. The wealth of successful semiempirical studies on organic reactions42 and the almost experimental accuracy of MM3 predictions for the geometries, relative energies, and heats of formation for many organic molecules43 document their relevance in this field. A hierarchy of three coupling models is presented in this paper. Model A is the simplest one and represents a mechanical embedding of the quantum region. Model B includes electrostatic interactions between the QM and MM regions using suitably defined classical point charges in the MM region which enter the core hamiltonian. The relaxation of the density matrix due to this external perturbation may be identified as the polarization of the QM region. The most refined model C also © 1996 American Chemical Society Quantum Mechanical and Molecular Mechanical Hybrid Models J. Phys. Chem., Vol. 100, No. 25, 1996 10581 includes the polarization of the MM region in the presence of the electric field generated by the QM region. Conceptually the coupling models focus on the theoretical description of the QM/MM interactions, without introducing any changes in the underlying QM and MM methods. This deliberate choice allows a rather general formulation that may also be applied to other QM/MM combinations. This paper is organized as follows: Section 2 deals with the general theory and includes a comparison of our models with earlier treatments. Section 3 describes the implementation of the models and briefly summarizes our semiempirical treatment of electrostatic interactions in models B and C which has been reported in detail elsewhere.44,45 Section 4 contains selected numerical applications, with a discussion of the merits and limitations of models A-C for particular problems. These applications include the calculation of heats of formation, proton affinities, and deprotonation enthalpies, as well as conformational analysis. Three types of organic reactions (hydride transfer, nucleophilic addition to carbonyls, and nucleophilic cleavage of oxiranes) serve as case studies which focus on the effects of substituents on activation energies. In the concluding section we report our experiences with the hybrid models and provide some general guidelines for future applications. total charge is not forced to be zero for neutral QM systems which might pose additional problems in the calculation of electrostatic QM/MM interactions. A simpler solution to these problems is to satisfy the free valencies with hydrogen atoms (“link atoms”, L) and to include these in the SCF treatment of the QM fragment. The use of link atoms corresponds to the chemically intuitive approximation of the charge density of a fragment as part of a molecule. Many ab initio or semiempirical studies take advantage of this concept when replacing e.g. large side chains by hydrogen atoms to define model systems which are feasible for QM calculations. In a hybrid QM/MM model, additional effects due to these omitted side chains are included in a perturbative fashion. The link atom approach has been adopted in several earlier QM/ MM models8,16 and will also be used here. In contrast to the earlier work, however, we start from a formula for the total energy which implicitly includes a molecular mechanical correction for the energetic contributions due to the link atoms. At the moment we assume that the MM energy expressions for X-Y and Y-L are well-defined quantities. In a QM/MM scheme, the total energy of the molecule X-Y may then be obtained from its MM energy by adding the QM energy for Y-L and subtracting the MM energy for Y-L (The superscript A in eq 2 refers to model A): 2. Theory A Etot (X-Y) ) EMM(X-Y) - EMM(Y-L) + EQM(Y-L) This section begins with a detailed discussion of the simplest model A (“Mechanical Embedding”). Special attention is paid to the treatment of QM/MM cuts within molecules where “link atoms” are used to satisfy free valencies at the QM/MM boundary. Models B (“Quantum Mechanical Treatment of Electrostatics”) and C (“Classical Treatment of Polarization”) are refined versions of the basic model A. They are only characterized by discussing the modifications relative to model A. 2.1 Model A: Mechanical Embedding. The combined QM/ MM description of a system composed of two separate molecules X and Y does not invoke any severe problems. One of the molecules (X) may be treated classically, and the other (Y) quantum mechanically, so that the development of a reasonable QM/MM model concentrates on a suitable representation of the intermolecular interaction energy E(X,Y). For the total energy Etot(X,Y) we obtain in an obvious notation: Etot(X,Y) ) EMM(X) + EQM(Y) + E(X,Y) (1) The present work, however, aims at a theoretical treatment of large organic molecules which are decomposed into MM and QM fragments. A suitable approach is much less evident in this case since there is no unambiguous quantum chemical treatment of a molecular fragment. None of the three obvious substitutes Y- (anion), Y• (radical), Y+ (cation) adequately represents the electron distribution of the fragment Y as part of the molecule X-Y. In the spirit of early work,7 Théry et al. have recently proposed a fragment orbital approach that involves a transformation of the atomic orbitals of the “frontier” QM atom yielding hybrid orbitals which are colinear with the bonds of this atom.10 Those hybrid orbitals which belong to bonds connecting QM and MM atoms are excluded from the orbital basis, and the associated electron densities act as external point charges on the cationic QM fragment. Although physically appealing, this approach suffers from the fact that the electron density of the excluded orbital is not known unless a QM calculation for the entire system is performed prior to the QM/MM treatment. The (2) EMM(X-Y) ) EMM(X) + EMM(X,Y) + EMM(Y) (3) EMM(Y-L) ) EMM(L) + EMM(Y,L) + EMM(Y) (4) Since the MM energy expression can unambiguously be decomposed into contributions from each of the two parts of the molecule and from their interaction (eqs 3, 4) one readily obtains a simplified formula which avoids the double evaluation of energy terms for Y A (X-Y) ) EMM(X) + EMM(X,Y) + Etot EQM(Y-L) + ELINK (5) ELINK ) -EMM(L) - EMM(Y,L) (6) ELINK denotes the link atom correction which will be discussed in more detail later. It should be kept in mind that the derivation of eq 5 is only formally valid, since the basic assumption of a well-defined expression EMM(X-Y) does not generally hold. Assuming that the fragment Y undergoes a chemical reaction, a molecular mechanical description is not possible, and EMM(Y) is ill-defined. Due to cancelation, however, this term does not appear in eq 5 which can therefore be regarded as a useful master equation. Similar problems may be encountered if the interaction terms EMM(X,Y) and EMM(Y,L) refer to atom types in Y for which no MM parameters are available. This concerns bonded (bond stretching, angle bending, torsion) interactions in the QM/MM overlap regions and nonbonded (electrostatic, van der Waals) interactions for the entire fragments X and Y. In practice, one either assumes reasonable parameters for the few missing bonded terms (which in many cases may be derived from similar bonding situations) and accepts nonbonded parameters which are generally available from either experimental data (van der Waals), other force fields (van der Waals, electrostatics), or quantum chemical calculations (mainly electrostatic), or one choses sufficiently large QM regions and thus avoids the problems associated with missing bonded parameters. Unlike most other force fields (e.g. UFF46 10582 J. Phys. Chem., Vol. 100, No. 25, 1996 Bakowies and Thiel (Y-L). It can be readily verified that minima of EQM(Y-L) with respect to link atom coordinates are found by minimizing A (X-Y) - ELINK. Consequently we define the QM/MM Etot potential energy by: A Epot (X-Y) ) EMM(X) + EMM(X,Y) + EQM(Y-L) (7) Figure 1. MM3 and MNDO C-H stretching potentials of methane. Starting from the MNDO geometry of methane, one hydrogen atom is moved along the C3 axis while all other atoms are fixed in their initial position. and those in AMBER,47-50 CHARMM51), MM3 includes many explicit couplings between bonded interactions (e.g. stretchbend) as well as far-reaching implicit couplings in π systems which are due to the readjustment of force field parameters using geometry-dependent π-SCF (VESCF52-54) density matrix elements.55 These couplings pose additional problems to the evaluation of EMM(X,Y), as discussed in detail elsewhere.44 Since they are expected to have only a marginal influence on the total energy and are completely absent in other commonly used force fields which are regarded as a future option for the proposed QM/MM approaches, we decided to neglect any MM3 coupling of diagonal terms involving internal coordinates in the QM region.44 With these specifications eq 5 provides a plausible and convenient definition of the total energy of a molecule. It still has to be examined, however, whether this energy formula is suitable for geometry optimizations. This is not self-evident since the formula implicitly includes the difference of the QM and MM interaction energies involving the link atoms L. A simple example demonstrates that this may cause problems. Figure 1 shows the QM (MNDO) and MM (MM3) stretching potentials for one of the CH bonds of the methane molecule, employing the MNDO geometry for the others and preserving C3V symmetry. While the MNDO and MM3 energy minima correspond to nearly the same CH bond lengths (1.104 and 1.112 Å, respectively), the energy curves are very different far off the minimum. The MM3 potential is much steeper left of the minimum, causing large discrepancies between MM3 and MNDO for shorter bond lengths. Since the corresponding energy difference is part of the QM/MM total energy A (X-Y) for a system composed of a QM fragment CH3, a Etot link atom H, and some MM fragment X, a QM/MM geometry optimization would continuously reduce the distance between the C atom and the link atom until they finally collapse. Geometry optimizations for several test cases show that this artifact is not restricted to stretching potentials but also occurs for other force field terms which refer to both QM and link atoms. In all these cases, this behavior is caused by large differences between the QM and MM potentials far away from the equilibrium reference geometry. Therefore, eq 5 cannot be used as a general definition of the QM/MM potential surface. Since both MM3 and MNDO have been calibrated against experimental data, the corresponding potential surfaces should be most similar in the vicinity of energy minima. Hence we may expect that ELINK (eq 6) is an appropriate correction to the link atom interactions implicitly included in EQM(Y-L) only for link atom positions which are close to the minimum of EQM- Conceptually this is equivalent to the definition of the potential energy and the total energy in the QM/MM model proposed by Field et al.8 In our treatment, the link atom correction ELINK is added to the potential energy EApot(X-Y), once the geometry is optimized. Such a procedure is not completely satisfactory from a theoretical point of view, since one would like to have potential energies and total energies which are the same. We have accepted this inconsistency in our treatment because ELINK appears to be an integral part of the total energy which corrects for some unphysical energy contributions associated with link atoms (see section 4 for further discussion). An alternative quantum mechanical link atom correction can easily be devised for MNDO type wavefunctions where the total energy consists of only one-center and two-center contributions.44 A plausible definition of a QM link atom correction would be the sum of all one-center and two-center contributions referring to link atoms. The resulting QM energy is, however, non-variational so that gradient evaluation becomes inefficient. Therefore, this approach was discarded in favor of the MM link atom correction discussed above. Model A is defined by eqs 5-7. We refer to it as mechanical embedding since all interactions between the QM and MM regions are included in EMM(X,Y) and are thus calculated molecular mechanically. 2.2 Model B: Quantum Mechanical Treatment of Electrostatics. MM3 (and its predecessor MM2) represent electrostatics by bond dipole interactions, contrary to many other force fields which employ partial atomic charges. Weaknesses of MM3 (and MM2) in the conformational analysis of molecules containing strongly polar groups56-58 are at least partly due to this approximation. A better representation of electrostatic QM/ MM interactions is an obvious goal for a refined hybrid model. This may already be achieved by simply replacing the bond dipoles with more appropriate potential-derived (PD)59 atomic point charges. In addition, these charges can explicitly be included in the hamiltonian for Y-L: Y X q J Ĥel(Y-L;X) ) Ĥel(Y-L) - ∑∑ i J riJ (8) Ĥel(Y-L;X) and Ĥel(Y-L) are the electronic hamiltonians for Y-L with and without the external field, respectively, which is generated by the MM atomic point charges qJ. It should be emphasized that Ĥel(Y-L;X) does not include any interactions between link and MM atoms, consistent with our basic assumptions (eq 5). In LCAO approximation, the inclusion of the MM charges changes the elements hµV of the core hamiltonian to X J h̃µV ) hµV - ∑qJVµV (9) J The resulting Coulomb or electrostatic energy is given by: Y Y X Y X J Ecoul(X,Y) ) ∑∑∑PµVqJVµV + ∑∑ZAqJVAJ µ V J A J (10) Quantum Mechanical and Molecular Mechanical Hybrid Models Here, PµV and ZA are density matrix elements and nuclear J and VAJ denote nuclear attraction charges, respectively. VµV integrals and Coulomb repulsion terms which describe the interaction between a unit charge at MM atom J and an electron or core A in the QM region, respectively. Using the modified core hamiltonian h̃ in the SCF procedure yields a perturbed density matrix P̃ and, consequently, a different SCF energy ẼQM(Y-L). This process may be identified as polarization or induction in the QM region and is accompanied by a net energy gain of Eind(Y) ) ẼQM(Y-L) - EQM(Y-L) - Ecoul(X,Y) (11) The refined model B includes the sum of the Coulomb and induction energies from eqs 10 and 11 which replace the electrostatic part of the QM/MM interactions from model A vdW (X,Y)) and bonded while keeping the van der Waals (EMM bonded (EMM (X,Y)) parts. The potential energy and the total energy are given by B (X-Y) ) EMM(X) + Ecoul(X,Y) + Eind(Y) + Epot (13) The idea to include MM atomic point charges in the core hamiltonian is not new.7-13 Apart from the link atom correction, model B is in fact conceptually analogous to several previous QM/MM approaches.8,9,11. We have adopted a different implementation, however, which aims at a realistic semiempirical description of electrostatic potentials for both the QM and MM regions of a molecule (see section 3 for a short summary and ref 45 for further details). 2.3 Model C: Classical Treatment of Polarization. Model B results in an unsatisfactory asymmetry for the description of nonbonded QM/MM interactions. While the polarization of the QM fragment is admitted, that of the MM fragment is not. Obviously, a term Eind(X) is missing (see eq 12) which describes the polarization of the MM fragment due to the presence of the QM electric field. A classical model frequently used in MD simulations of ionic systems (see, e.g., ref 60) treats the induction energy as a sum of atomic contributions: 1X Eind(X) ) - ∑µJR⟨FJR⟩ 2 J (14) and thus accounts for the nonuniform distribution of the electric field.44 (The notation in section 2.3 as introduced in eq 14 implies the summation over repeated Greek indices.) In the context of our QM/MM model the electric field may be identified with the expectation value of the unit field operator. In LCAO approximation, the corresponding R component for location J reads as follows: Y Y ⟨FJR⟩ ) ∑∑ µ V Y J P̃µV∇RVµV - ∑ZA∇RVAJ ever, as was already recognized in 1917 by Silberstein.61 In the spirit of this early work, Applequist proposed an induced dipole interaction model62 which, however, suffered from resonance problems for dipoles close in space (e.g. in a chemical bond). The parametrization of his model consequently yielded atomic polarizabilities which were much too small compared to the available experimental and theoretical (ab initio) data and showed the need to differentiate between identical atoms in dissimilar chemical environments. Later, Thole proposed a revised model which in an empirical fashion took into account the physically well-known damping behavior of short-range polarization.63 His model outperforms the older one in accuracy although it only needs one parameter per element, the isotropic atomic polarizability RJ. The calibration of his model against observable molecular polarizabilities yields RJ values which are in much closer agreement with experimentally available or calculated (high level ab initio) polarizabilities of free atoms, indicating the physically adequate description of short-range damping behavior.44 According to Thole’s model the induced dipole moment of atom I is given as:63 X bonded vdW (X,Y) + EMM (X,Y) (12) EQM(Y-L) + EMM B B (X-Y) ) Epot (X-Y) + ELINK Etot J. Phys. Chem., Vol. 100, No. 25, 1996 10583 (15) A The atomic dipole moments µJR still need to be defined. The simple assumption that they are independently induced by the external field (µJR ) RJ⟨FJR⟩) would require strict additivity of atomic polarizabilities (RX ) ∑JXRJ). This is not true, how- IJ J µIR ) RI(⟨FIR⟩ + ∑tRβ µβ) (16) IJ IJ ) ν4TRβ + 4(ν4 - ν3)rIJ-3δRβ tRβ (17) J*I IJ TRβ denotes the usual dipole-dipole interaction tensor which may conveniently be defined as the second derivative IJ ) ∇R∇βrIJ-1), matrix of the inverse interatomic distance (TRβ δRβ is the Kronecker delta, and ν is a scaling function which mimics the damping behavior (ν ) rIJ/sIJ if rIJ < sIJ and ν ) 1 otherwise, where sIJ ) 1.662(RIRJ)1/6 63). A physically reasonable and well-balanced model of induced dipole interaction can thus reproduce molecular polarizabilities from measurable atomic quantities. This observation provides empirical evidence for the assumption that the interatomic polarization is only a higher order correction to otherwise additive polarizabilities. It has indeed only recently been shown that the classical model of induced dipoles (or multipoles, in general) which mutually polarize themselves until selfconsistency is achieved, corresponds to infinite-order intermolecular perturbation theory while the second-order expression only includes the polarization due to the external field.64 Therefore it might be tempting to adopt the simple approximation of additive atomic polarizabilities. On the other hand, the induced dipole interaction model is now well-established in classical MD simulations although usually in the older, less satisfactory implementation of Applequist. More importantly, it represents the only way to describe molecular anisotropy using simple scalar parameters, i.e. isotropic atomic polarizabilities. Thole’s approach performs very satisfactory in this respect, yielding polarizability tensor components which are usually accurate to within 6%.63 From a theoretical point of view the induced dipole interaction model is supported by the observation that it treats the MM fragment in closer analogy to the quantum mechanical description of the QM fragment. The SCF procedure obviously includes all orders of intermolecular perturbation theory and thus accounts for the “internal polarization”. However, a classical treatment of polarization formally equivalent to the QM (SCF) description would also include nonlinear effects which are part of the third and higher order expressions of intermolecular perturbation theory.64,65 A preliminary study on the water dimer (one molecule treated by QM, the other by MM) has indicated that the nonlinear contributions to the QM induction energy are negligible at the semiempirical level.44 10584 J. Phys. Chem., Vol. 100, No. 25, 1996 Bakowies and Thiel We adopt the induced dipole interaction model as specified by eqs 14-17 to describe the polarization of MM fragments by QM electric fields, and define model C by charges and point polarizabilities without referring to a molecular mechanical treatment of the classical region. 3. Implementation C Epot (X-Y) ) B Epot (X-Y) + E (X) ind C C (X-Y) ) Epot (X-Y) + ELINK Etot (18) (19) Test calculations using model C indicated an unreasonably large polarization of MM fragments, due to the lack of a proper damping mechanism for MM atoms participating in QM-MM bonds. We remedied this artifact by simply treating all such MM atoms at the QM/MM boundary as unpolarizable. This is a very crude assumption, but one may argue that the major part of the short range polarization is experienced by MM atoms directly connected to the QM fragment and that these effects are already implicitly included in the QM treatment of the link atom which acts as a substitute. Nevertheless, future developments are likely to focus on a refinement of our treatment in this respect. The electric field ⟨FJR⟩ is evaluated using the perturbed density matrix P̃ which refers to the modified core hamiltonian h̃ (see eq 9). This procedure avoids the need for a second SCF calculation using the initial core hamiltonian h. In this way, induction energy terms referring to a third-order treatment of the “interfragment” polarization are included.44 Contributions from higher orders of intermolecular perturbation theory may be included in a reaction field model which describes the mutual polarization of QM and MM fragments. Two different types of reaction field hamiltonians are known in the literature, describing an average (ARF)66 and a direct (DRF)67 reaction field, respectively, the latter referring to an instantaneous response. There is some discussion about the physical implications of either approach,68 but the ARF hamiltonian seems to model the reaction field more appropriately.69 We tested an implementation analogous to the ARF model introduced by Tapia and Goscinski,66 with the difference that the response is generated by a discrete reaction field (atomic polarizabilities) rather than a continuum.44 Not too surprisingly, the effects on the total induction energy were negligible, since model C already includes all second-order and some third-order contributions.44 In an independent study which appeared recently,12 Thompson and Schenter presented a QM/MM model conceptually almost identical with our reaction field implementation. Their main focus was the investigation of excited state properties of the bacteriochlorophyll D dimer, and the inclusion of MM polarization appeared to be necessary for a satisfactory agreement with Stark effect experiments. Unfortunately, no results are reported for a simpler polarization treatment analogous to our model C. According to our experiences, the additional effort of a self-consistent reaction field is not justified at the semiempirical level, since only marginal differences in energy were observed in any of the test calculations. Hence we did not further study the reaction field model but decided to accept model C as our most refined QM/MM approach. Apart from the recent work of Thompson and Schenter,12 there are two earlier QM/MM approaches which include approximate treatments of polarization.7,16 The more recent approaches, however, did not take up this feature. The physically most appealing polarizability model of Thole63 has so far only been considered in the DRF approach of van Duijnen and co-workers67,70 which is, however, not a hybrid QM/MM approach in the narrow sense, since it is mainly concerned with a QM description of a molecule in the field of classical point A computer program called MNDO/MM has been written on the basis of MNDO9171 and MM3(89)72 which allows practical applications of the proposed QM/MM models using either MNDO37 or AM139 wavefunctions and the MM3 force field.40,41 Details of the implementation are given elsewhere;44,45 here we only summarize the most important aspects which include the semiempirical evaluation of electrostatic and induction interactions and the computation of energy gradients. The adequate treatment of electrostatic QM/MM interactions is the main focus for a proper implementation of models B and C. Using eq 10 we may rewrite the Coulomb interaction between QM and MM fragments as X Ecoul(X,Y) ) ∑qJΦJ (20) J where ΦJ denotes the QM electrostatic potential at site J: Y Y Y V A J ΦJ ) -∑∑PµVVµV + ∑ZAVAJ µ (21) The electrostatic potential is a well-defined quantity only in ab initio quantum chemistry. In semiempirical methodology, J and VAJ. The there are several options for the evaluation of VµV 73-76 which is characterized by “quasi ab initio” approach J and VAJ employing deorthogoanalytic evaluation of both VµV nalized semiempirical wavefunctions has become popular for many applications including the calculation of potential derived charges.77,78 It is, however, less satisfactory in the context of QM/MM approaches, since the deorthogonalization procedure is very time-consuming and affords additional three center integrals (VµJ AVB) which make the approach even less efficient. We adopted a parametrized approach which has first been proposed by Ford and Wang79 and assumes an orthogonal basis in line with the basic NDDO assumptions:45 Both quantities, J and VAJ, are represented by MNDO type parametric VµV functions which have been calibrated against ab initio RHF/631G* electrostatic potentials of a typical set of small organic and inorganic molecules. Compared to the original parametrization of Ford and Wang79 which was mainly concerned with minima of electrostatic potentials, we used a larger set of reference points on several molecular surfaces with distances ranging from 0.6 to 1.2 times the van der Waals radii and also included electric field components in some of the parametrizations.45 This admitted a more balanced calibration of electrostatic potentials and electric fields for regions close to and far away from the molecules. The MM atomic point charges qJ (eq 20) are derived from a charge equilibration scheme (QEq)45,80 which is based on the principle of electronegativity equalization. The original QEq approach of Rappé and Goddard80 has been adapted to our semiempirical QM/MM methodology, affording KlopmanOhno type two center integrals which converge to adjustable atomic integrals in the one-center limit.45 This semiempirical QEq approach involves two parameters per element which have been calibrated to reproduce ab initio RHF/6-31G* potential derived (PD) or Mulliken charges.45 In the present QM/MM study we employ the one-parametric function f 1′ defined in eq 10a of ref 45 to calculate electrostatic potentials and use the PD charge parametrization for the QEq Quantum Mechanical and Molecular Mechanical Hybrid Models charge equilibration scheme. According to previous experience, an accuracy of about (20-25% and (0.1 e should be expected for potentials and charges, respectively, when compared to ab initio RHF/6-31G* reference data.45 Analytic energy gradients are readily available for model A by simply combining the QM and MM gradients. In model B, the inclusion of MM point charges in the core hamiltonian matrix generates additional forces on QM (gxA) and MM (gxJ) atoms: X ∂VAJ J ∂xA gxA ) ∑ZAqJ Y ∂VAJ A ∂xJ gxJ ) ∑ZAqJ X A A ∂VµJ AVA J µA VA ∂xA - ∑∑∑P̃µAVAqJ Y A A ∂VµJ AVA A µA VA ∂xJ - ∑∑∑P̃µAVAqJ (22) + g′xJ (23) J. Phys. Chem., Vol. 100, No. 25, 1996 10585 and electric field strengths (⟨F1x⟩, ⟨F1y⟩, ⟨F1z ⟩, ⟨F2x⟩, ..., ⟨Fnz ⟩), respectively. The elements of the supermatrix A are given as: JJ ARβ ) (RJ)-1δRβ (27) IJ IJ ) -TRβ ARβ (28) Using eqs 26-28, the MM induction energy may be evaluated as 1 Eind(X) ) - ⟨F+⟩A-1⟨F⟩ 2 (29) The matrix inversion technique is numerically stable but much slower than the iterative procedure, typically by factors of 5-10. Therefore, the latter has been preferred whenever possible. 4. Numerical Applications The total QM energy expression (EQM(Y-L) + Ecoul(X,Y) + Eind(Y)) is stationary with respect to the perturbed density matrix P̃ so that the QM gradient expressions do not involve any density matrix derivatives.81 The QEq scheme yields charges which depend on the geometry of the MM fragment. This leads to additional forces g′xJ exerted on MM atoms: Y Y A A ∂qJ ∂qJ g′xJ ) ∑ZA VAJ - ∑∑∑P̃µAVA VµJ AVA ∂xJ ∂xJ A A µA VA (24) The charge derivatives ∂qJ/∂xJ in eq 24 are readily available.45 No analytic energy gradients are provided for model C since the QM energy expression is nonvariational in this case. A proper implementation would require the solution of CPHF type equations, a time-consuming procedure which seems inadequate in the context of a simple QM/MM approach. Test calculations have shown, however, that the additional consideration of polarization in the MM fragment mainly affects the QM/MM total energy, with little or no effect on the molecular geometry.44 Some examples are discussed in the next section which justify the single point evaluation of model C energies at geometries which have been optimized using model B. We report these results in analogy to common notation as C//B (total energy// geometry). The evaluation of MM induction energies Eind(X) deserves a final comment. The iterative procedure defined by eqs 14-17 may converge slowly or even diverge, especially when charged QM fragments generate large electric fields. The convergence behavior can be much improved by introducing a damping mechanism µJR(new): ) (1 + f)µJR(new) + fµJR(old) (25) Applying damping factors f in the range of 0.4 to 0.6 usually leads to satisfactory convergence ((10-5 Debye) after 20 iterations. A matrix inversion technique62,63 may be used if the iterative procedure does not converge at all. For this purpose eq 16 may be rewritten in matrix notation: M ) A-1⟨F⟩ (26) M and ⟨F⟩ denote column vectors which collect the compo) , µind,1 , µind,2 , ..., µind,n , µind,1 nents of dipole moments (µind,1 z x z x y Selected numerical applications have been studied to establish the accuracy of the proposed QM/MM models and their ability to reproduce experimental trends reliably. In particular, we have compared the performance of models A-C to assess the appropriate levels of QM/MM embedding required for different types of application. The QM/MM boundary has often deliberately been put close to the reactive center to emphasize electrostatic and related effects, and different choices of QM/ MM boundaries have been investigated to derive guidelines for future applications. Although the discussion in this section will stress such methodological issues, most of the examples chosen are also relevant from a chemical point of view and therefore illustrate the potential of QM/MM applications. 4.1 Heats of Formation of Organic Molecules. MNDO (AM1) and MM3 are designed to calculate heats of formation. It is straightforward to convert QM/MM total energies to heats of formation, using bond and structure heat increments for the MM part of the energy expression, and atomic energies and atomic heats of formation for the QM part.44 Figure 2 shows the heat of formation of 1-octanol calculated for various QM/ MM divisions, treating the hydroxyl group either molecular mechanically (top chart) or quantum mechanically (bottom chart). The QM/MM results are of the same order as the MNDO and MM3 values, with maximum deviations of ≈5 kcal/mol (top) or ≈10 kcal/mol (bottom). These deviations are almost entirely due to the differences between MNDO and MM3 for the QM part of the molecule. For example, the deviation between the MNDO and MM3 values for methanol to 1-heptanol decreases from -9.6 to -2.0 kcal/mol, in analogy to the QM/MM results in the bottom chart. Apart from one single exception, the heats of formation from model B are consistently lower than those from model A, by 2 to 4 kcal/mol (see Figure 2), while the inclusion of MM polarization in model C has only minor effects (less than 0.4 kcal/mol, not shown). The above and other examples44 indicate that it is possible to define a heat of formation in our QM/MM treatment and to obtain reasonable results which are not overly dependent on the choice for the QM/MM boundary. However, there are several reasons to avoid the concept of a heat of formation in our QM/MM models. MNDO (AM1) considers the heat of formation as a conformational property which characterizes distinct stationary points, while MM3 defines it as a molecular property which in general requires the consideration of torsional and population increments. Since the quality of MM3 heats of formation largely depends on the careful calibration of heat increments, meaningful QM/MM results would be restricted to 10586 J. Phys. Chem., Vol. 100, No. 25, 1996 Bakowies and Thiel TABLE 1: Relative Energies of Various Ethylene Glycol and Ethylenediamine Conformationsa,b MNDO/MM3d conformationc MNDO MM3 A//A ab initioe B//B C//C RHF MP2 0.0 4.9 9.7 12.5 0.0 5.4 10.8 15.7 0.0 4.8 10.4 15.2 0.0 2.4 3.6 12.8 0.0 2.8 6.0 14.8 0.0 3.8 6.0 15.8 C2h (a) C2h (s) C2V (a) C2V (s) 0.0 4.1 3.1 9.1 HOCH2CH2OH 0.0 0.0 0.0 4.7 4.3 4.9 7.9 8.0 9.7 12.9 12.4 12.7 C2h (a) C2h (s) C2V (a) C2V (s) 0.0 0.2 3.1 5.5 H2NCH2CH2NH2 0.0 0.0 0.0 3.1 3.1 2.3 4.6 4.7 3.6 9.8 10.0 12.7 a All energies are in kcal/mol. b All methods predict a minimum and a saddle point of first, second, and third order for the C2h (a), C2V (a), C2h (s), and C2V (s) conformations of both molecules, respectively. c (a) antiperiplanar, (s) synperiplanar conformation, with respect to the C-O and C-N bonds, respectively. d QM(XCH2)/MM(CH2X). e 6-31G* basis set. Figure 2. MNDO/MM3 heats of formation of 1-octanol. cases tractable by MM3 alone. Furthermore, the MM3 heat increments have been derived in a manner consistent with the MM3 treatment of electrostatics, so that they would not be strictly applicable to models B and C with their own refined treatment of QM/MM electrostatic interactions. In view of such inconsistencies, we shall not refer to heats of formation in the following subsections, but report QM/MM energies as defined in eqs 5, 7, 12, 13, 18, and 19. 4.2 Conformational Analysis. Two molecules, ethylene glycol and ethylenediamine, have been chosen to illustrate the performance of the QM/MM models in conformational analysis. Four symmetrical (C2V and C2h) conformations have been fully optimized applying all three QM/MM models, MNDO, MM3, as well as the RHF/6-31G*1,82 and MP2/6-31G*1,82 levels of theory for reference. In the QM/MM calculations, the molecules have been divided at the central CC bond, thereby lowering the symmetry to Cs. Since MNDO/MM3 and AM1/MM3 yield very similar results, only the first are shown and discussed. All relevant data are collected in Table 1. Apart from MNDO, all methods predict the same sequence in energies. Not too surprisingly, MM3 and MNDO/MM3/A (denoting a QM/MM calculation at the model A level, employing MNDO and MM3) yield very similar results. Both are much more accurate than MNDO when compared to the ab initio results as a reference. The quantum chemical treatment of Coulomb interactions between the fragments improves the QM/ MM description for ethylene glycol, and for the least stable conformation of ethylene diamine. The results from models B and C are very similar, indicating only a small influence of polarization effects on the relative energies. The geometry optimizations using models B and C lead to very small differences in bond lengths (e0.002 Å), bond angles (e0.3 degrees), and dihedral angles (e0.3 degrees). C//B and C//C energies differ by less than 0.01 kcal/mol. These and other analogous results justify the use of geometries from model B for energy evaluations using model C. 4.3 Protonations and Deprotonations. Two elementary reactions with high relevance for organic chemistry and biochemistry are protonations and deprotonations of Lewis bases and acids, respectively. Due to large differences between the charges of reactants and products, substituent effects of 10 kcal/ mol and more are common in these processes. We studied the protonation of alcohols and pyridines as well as the deprotonation of alcohols and carboxylic acids.44 Here we report the results for the alcohols and discuss the ability of the QM/MM approaches to model the electrostatic and polarization effects of alkyl substituents in these reactions. The proton affinity PA of a base B and the deprotonation enthalpy DPE of an acid HB are thermodynamically defined as PA ) ∆Hf0(H+) + ∆Hf0(B) - ∆Hf0(HB+) (30) DPE ) ∆Hf0(H+) + ∆Hf0(B-) - ∆Hf0(HB) (31) Since the heat of formation ∆Hf0 of the proton is only poorly reproduced by MNDO type methods, the experimental value is normally used in semiempirical studies.83,84 We have proceeded accordingly, with ∆Hf0(H+) ) 365.7 kcal/mol (“stationary electron convention”).85 Several QM/MM divisions have been tested, but the protonated or deprotonated part of the molecule has always been treated quantum mechanically. Bonds which are formed by protonation of anions or of neutral molecules are treated as normal bonds in the molecular mechanical part of the QM/MM coupling term. All quoted results refer to the energetically most favorable conformations which may, however, be different for the various theoretical approaches (see supporting information). The QM/MM results generally refer to C//B calculations if not stated otherwise. Proton Affinities of Alcohols. The proton affinities of oxygen compounds as electron donors generally increase with successive methyl substitution. In most experimental86-89 and theoretical studies90 this phenomenon is attributed to inductive91 (charge transfer) and polarization effects. The relative significance of these and other factors such as electrostatics, however, strongly depends on the type of correlation of experimental data Quantum Mechanical and Molecular Mechanical Hybrid Models J. Phys. Chem., Vol. 100, No. 25, 1996 10587 TABLE 2: Proton Affinities of Alcohols: Relative Valuesa AM1(CH3OH)/MM3 molecule RR Rβ Rγ expb AM1 C//B R3COH R3COH R3COH R3COH R3COH H CH3 C2H5 CH3 CH3 H H H CH3 CH3 H H H H CH3 0.0 6.4 8.9 9.3 11.8 0.0 6.2 7.3 10.8 15.9 0.0 7.7 8.3 13.0 17.4 AM1(H2O)/MM3 B//B B Epot A//A C//B B//B B Epot 0.0 5.3 5.2 8.0 9.7 0.0 3.9 3.7 5.5 5.7 0.0 0.8 1.2 1.8 1.6 0.0 0.3 1.6 -0.9 -1.8 0.0 -0.7 -0.7 -2.3 -3.0 0.0 -0.9 -0.8 -2.5 -3.4 a Absolute values for methanol are: 181.9 (exp), 170.4 (AM1 and AM1(CH OH)/MM3), 176.2 (AM1(H O)/MM3/C//B). All entries are in 3 2 kcal/mol. b See: Lias, S. G.; Liebman, J. F.; Levin, R. D. J. Phys. Chem. Ref. Data 1984, 13, 695. TABLE 3: Deprotonation Enthalpies of Alcohols: Relative Valuesa AM1(CH3OH)/MM3 AM1(H2O)/MM3 molecule RR Rβ Rγ expb AM1 C//B B//B B Epot C//B B//B B Epot R3COH R3COH R3COH R3COH R3COH R3COH R3COH R3COH R3COH R3COH H CH3 C2H5 t-C4H9 CH3 CH3 C2H5 i-C3H7 t-C4H9 CH3 H H H H CH3 t-C4H9 t-C4H9 t-C4H9 t-C4H9 CH3 H H H H H H H H H CH3 0.0 -3.1 -4.5 -7.1 -5.1 -8.5 -9.6 -10.7 -11.9 -5.9 0.0 0.2 -1.1 -3.3 0.3 -2.8 -3.8 -4.4 -5.3 -1.2 0.0 0.6 -0.3 -1.3 -0.9 -1.6 -2.3 -3.3 -3.6 -8.2 0.0 1.0 0.8 2.4 -1.6 0.1 0.7 1.0 1.8 -8.6 0.0 2.3 2.0 3.5 1.1 2.4 3.0 3.2 3.7 -4.0 0.0 -4.6 -6.4 -6.8 -9.2 -11.9 -12.9 -15.6 -16.6 -13.3 0.0 -2.0 -2.3 -0.5 -3.9 -2.6 -2.2 -2.5 -2.4 -5.7 0.0 -2.0 -2.3 -0.4 -3.9 -2.6 -2.2 -2.5 -2.5 -5.7 a Absolute values for methanol are: 379.2 (exp), 384.2 (AM1 and AM1(CH3OH)/MM3), and 417.7 (AM1(H2O)/MM3/C//B). All entries are in kcal/mol. b From ion cyclotron resonance experiments ((2 kcal/mol). See Bartmess, J. E.; Scott, J. A.; McIver, R. T., Jr. J. Am. Chem. Soc. 1979, 101, 6046. or the particular energy partitioning scheme applied in theoretical work. Table 2 lists our results for some simple alcohols. It is obvious that AM1 reproduces the trends in the experimental proton affinities reasonably well. In the AM1(CH3OH)/MM3 partitioning, the mechanical embedding of model A yields only very small changes of the proton affinities and thus fails qualitatively which may be taken as indirect evidence for the importance of electrostatic effects. The simple electrostatic embedding of model B indeed accounts almost quantitatively for the observed increase in the proton affinities upon adding the first, second, and third methyl group to methanol (exp 6.4, 2.9, and 2.5 kcal/mol; calcd 5.3, 2.7, and 1.7 kcal/mol). The dominant differential stabilization of the acid relative to the conjugate base is provided by the electrostatic interactions in model B (e.g. in the case of ethanol: 4.6 kcal/mol out of a total of 5.3 kcal/mol). Model C includes the attractive induction interaction in the MM part which favors the charged species and increases with the size and the chain length of the polarizable MM fragment. Consequently, the results from model C further emphasize the increase of the proton affinities with successive methyl substitution, and they give the correct trend for extending the chain length (contrary to model B, see ethanol vs 1-propanol). Table 2 also shows the results for the AM1 (H2O)/MM3 partitioning which are clearly unsatisfactory. In this case, the QM/MM boundary divides the polar CO bond directly adjacent to the reactive center (i.e. the oxygen atom), and it is therefore not too surprising that the electronic rearrangements during protonation cannot be fully represented by the simple QM/MM interactions included in our models. It is apparently more appropriate to apply the QM/MM approach when the QM reactive center and the MM fragment are separated by two bonds, as e.g. in the AM1(CH3OH)/MM3 calculations. Deprotonation Enthalpies of Alcohols. The deprotonation enthalpies of alcohols decrease with successive alkyl substitution and with extended chain lengths (see Table 3). AM1 severely underestimates these substitution effects, but normally predicts their sign correctly. In the AM1(CH3OH)/MM3 partitioning, the inclusion of MM polarization in model C is necessary for obtaining the correct sign, whereas model B fails in this regard (see Table 3). The AM1(H2O)/MM3 calculations with model C yield numerical results rather close to the experimental values which must at least partly be due to fortuitous error compensation (considering that the QM/MM boundary cuts the CO bond, see above). For both partitionings, however, model C provides a significant improvement over model B indicating, in our opinion, that model C captures an essential physical effect: The indcuction energy increases with the polarizable volume of the alkyl rest and preferentially stabilizes the anions whose rather localized charges generate large electric fields in the alkyl regions. There are some interesting parallels between our computational results and empirical understanding.87,88 The first experiments have been conducted in solution phase and yielded the acidity sequence CH3OH > C2H5OH > t-C4H9OH.92 Subsequent gas phase experiments, however, revealed a reversed acidity sequence87,88 which could only be rationalized with strong polarization favoring large anions.88,89,93 ”Thus, the solution order is probably an artifact, and does not represent any intrinsic property of the molecule.” 88 More recent gas phase NMR studies on alcohols provided some evidence that electronic factors affecting the neutral species might also be responsible for the observed acidity sequence.94 The results did not admit, however, any quantitative estimate of these effects. Discussing their own QM/MM model, Field, Bash, and Karplus attribute the large errors of AM1(H2O)/MM deprotonation enthalpies for alcohols to the poor AM1 results for small anions (OH-).8 This is certainly not wrong, but the qualitatively incorrect increase in deprotonation enthalpy from H2O to CH3OH which is characteristic of their (+10.5 kcal/mol)8 and our (C//B: +6.9 kcal/mol, compare to AM1: -26.6 kcal/mol, exp: -11.6 kcal/mol, see supporting information) result, is merely caused by the exaggerated Coulomb repulsion between the MM methyl fragment and the directly connected QM 10588 J. Phys. Chem., Vol. 100, No. 25, 1996 Bakowies and Thiel Figure 4. QM/MM treatment of hydride transfer reactions. According to option a, only the reactive center is treated quantum mechanically. In option b, the classical region is very small and only consists of the hydrocarbon chains connecting the bridgeheads of molecules 1-3. TABLE 4: Activation Energies of Hydride Transfer Reactionsa AM1/MM3/A Figure 3. Transition structures of hydride transfer reactions. Symmetry point groups: Cs (1-4) and C2V (5). reactive center. Again, this illustrates the problems of having the QM/MM boundary too close to the reactive center. Further Remarks. Tables 2 and 3 also report proton affinities and deprotonation enthalpies obtained using AM1/MM3/B//B potential energies (EBpot(X-Y)). Obviously, the link atom correction is significant only when CH3OH is treated quantum mechanically. In this case, however, it leads to a considerable improvement of the results, especially for proton affinities. Our discussion has been restricted to the AM1/MM3 results, since MNDO/MM3 calculations do not show any qualitative differences (see supporting information). All model C calculations have also been performed using fully optimized (C//C) instead of model B geometries (see supporting information). The total energies normally differed by no more than 0.1, 0.4, and 1.0 kcal/mol for neutral, deprotonated, and protonated molecules, respectively, except for t-C4H9OH (3 QM/MM divisions; deviations: 0.2, 0.9, and 2.0 kcal/mol, respectively). None of the qualitative conclusions is affected by the “accuracy” of the applied geometries. 4.4 Hydride Transfer Reactions. The symmetric hydride transfer of deprotonated hydroxy ketones 1-5 (see Figure 3) has been studied kinetically by dynamic NMR spectroscopy.95-97 Watt and co-workers aimed at an experimental access to the reaction path of Meerwein-Ponndorf-Verley-Oppenauer redox cycles by correlating characteristic geometrical parameters of the sterically congested molecules with measured activation barriers. Since the geometry of the reactive center is nearly completely determined by the rigid hydrocarbon backbones, this reaction provides an ideal example for mechanical embedding. No significant electrostatic effects are expected to influence the reaction, hence the simplest QM/MM model A should be able to model the reaction satisfactorily.98 Here we summarize the main results of our AM1/MM399 study on these hydride transfer reactions and analyze the ability of model A to reproduce trends in activation energies and transition state geometries. The results are compared to recent TSM100 (transition state modeling) calculations,101,102 and the relation between TSM and QM/MM models is discussed. A full report of our study is given elsewhere.44 It includes a discussion of the reactant geometries and the effects of alkyl substituents on activation energies, provides additional examples, and also addresses correlations between geometries and activation energies. Figure 4 illustrates the two different QM/MM divisions which have been chosen for the calculations. According to option a, only the reactive center (CH2O + H-+ CH2O) is treated molecule option a 1 2 3 4 5 11.7 9.5 7.9 4.1 10.9 option b AM1 MP2/6-31+G*b expc 18.0 15.6 14.0 d d 18.1 16.8 15.7 7.3 12.5 16.5 13.0 11.4 2.9 6.6 21.7 (1.9) 19.0 (2.0) 17.3 (2.1) 13.7 (2.1) 19.4 (0.3) a All entries in kcal/mol. b MP2/6-31+G*//AM1. c See refs 95-97. The experimental errors are estimated to be (0.2 kcal/mol (3 to 5) and (0.3 kcal/mol (2), respectively. The experimental value for 1 is reported as a lower limit. Computed corrections to convert from measured quantities to activation energies at 0 K are given in parentheses (see text). d Option b is only defined for molecules 1-3. quantum mechanically, while for the alternative option b, the classical region comprises the hydrocarbon bridges (CH(CH2)nCH) present in molecules 1 to 3. The entire reaction path has been modeled with just one predefined set of atom types and bonds such that the number and type of force field terms entering the MM part of the QM/MM coupling term remains the same for reactants, transition states, and products. The symmetry of the reaction requires that the hydride ion has no MM bonds to any of the carbon atoms. Furthermore, the carbonyl and alkoxy groups need to be represented by the same MM parameters. The use of alkoxy parameters for both groups tends to underestimate the rigidity of the reaction center. Employing carbonyl parameters is not ideal either but prevents unreasonable geometries. The best solution would be provided by a new calibration, but we decided to use carbonyl parameters to avoid any parameter proliferation at this early stage of testing the QM/ MM model. To provide sufficient reference data for comparison, we supplemented the available experimental results with AM1 and ab initio calculations.82 Small model systems were used to check the accuracy of AM1 and affordable ab initio levels against MP2 calculations with large basis sets (up to triple ζ quality, and augmented with diffuse and polarization functions).44 Surprisingly, AM1 turned out to predict geometries more reliably than RHF/3-21G and RHF/3-21+G.44 For activation energies, the most adequate and still feasible level was MP2/ 6-31+G*//AM1.44 Table 4 collects the AM1/MM3/A//A activation energies, supplemented with theoretical (AM1, MP2/6-31+G*) and experimental reference data. The latter are Gibbs free enthalpies (∆Gqexp) and thus require conversion to activation energies (∆Eq) for a meaningful comparison. Appropriate corrections have been estimated from RHF/3-21G geometries and scaled (0.9) vibrational frequencies using the harmonic oscillator and rigid rotor approximation without explicit treatment of low- Quantum Mechanical and Molecular Mechanical Hybrid Models J. Phys. Chem., Vol. 100, No. 25, 1996 10589 TABLE 5: Relative Activation Energies Of Hydride Transfer Reactionsa AM1/MM3/A molecule 1 2 3 4 5 option option a b TSMb AM1 MP2/6-31+G*c expd 0.0 -2.2 -3.8 -7.6 -0.8 0.0 -1.3 -2.4 -10.8 -5.6 0.0 -3.5 -5.1 -13.6 -9.9 0.0 -2.6 -4.2 -7.8 -3.9 0.0 -2.4 -4.0 e e 0.0 -2.0 -3.4 -6.2 2.3 a All entries in kcal/mol. b Transition state modeling results from ref 101. c MP2/6-31+G*//AM1. d Corrected experimental values, see Table 4. e Option b is only defined for molecules 1-3. TABLE 6: Nonbonded Distances in the Transition Structures (in Å) AM1/MM3/A distance structure option a C‚‚‚Ca 1 2 3 4 5 1 2 3 4 5 2.495 2.465 2.443 2.528 2.749 1.448 1.434 1.425 1.393 1.444 C‚‚‚Hb option b AM1 2.412 2.387 2.369 c c 1.458 1.443 1.432 c c 2.411 2.390 2.373 2.425 2.676 1.448 1.436 1.428 1.373 1.418 a Distance between the C atoms of the CO groups. b Distance between the migrating H atom and the C atom of a CO group. c Option b is only defined for molecules 1-3. frequency vibrations and neglecting electronic contributions.44 Applying these corrections, the experimental values appear to be 7-13 kcal/mol higher than the best available ab initio data. This discrepancy may partly be attributed to solvation effects. Furthermore, the experimental values may not be accurate103 since the reaction rates strongly depend on substrate concentration and counterions.104 Apparently, the discrepancy is much smaller for molecules 1-3 than for molecules 4 and 5. Consequently, there is a good agreement between theory and experiment (corrected values) for relatiVe activation energies only within the groups (1, 2, 3) and (4, 5) (see Table 5). Facing the uncertainties in the experimental values and admitting that the theoretical level is far from being definitive, no decision can be made as to which set of reference data is most reliable. Higher level ab initio calculations are certainly desirable, but appear to be prohibitive currently. AM1/MM3/A (option b) predicts absolute activation energies close to the AM1 values. This is not surprising at all, since the QM region is very large in this case. The length of the hydrocarbon chain (treated molecular mechanically) affects the reactive center only indirectly through adjusting its geometry at the bridgehead hinges. Interestingly, AM1/MM3/A (option b) yields relatiVe activation energies in closer agreement than AM1 with both experimental and ab initio reference data. If just the reactive center is treated quantum mechanically (option a), AM1/MM3/A predicts relatiVe activation energies in nearly perfect agreement with the experimental values for 1-4 (see Table 5), with differences of no more than 0.4 kcal/mol, a value characteristic of the experimental error limits. Even the result for 5 seems to be acceptable, given the discrepancies observed for the other theoretical approaches. AM1/MM3/A (option a) underestimates, however, the absolute activation energies. Selected geometrical parameters characterizing the transition structures are collected in Table 6. As expected, AM1/MM3/A predicts nonbonded C‚‚‚C and C‚‚‚H distances at the reactive Figure 5. Decomposition analysis of the AM1/MM3/A (option a) activation energies of hydride transfer reactions. TOTAL, QM, and LINK denote contributions from the QM/MM total energy A (X-Y), the QM energy for the QM subsystem ∆EQM(Y-L) and ∆Etot the link atom correction ∆ELINK, respectively. REST denotes the A difference between ∆Etot (X-Y) and the sum of ∆EQM(Y-L) and ∆ELINK. center in close agreement with AM1 when only the hydrocarbon chains are treated classically (option b). AM1/MM3/A thus successfully models the mechanical compression of the reactive center geometry due to extending MM hydrocarbon chains which open the bridge heads. Compared to AM1, the QM/MM model yields C‚‚‚C distances which are consistently larger (by 0.07-0.10 Å) when only the reactive center is treated quantum mechanically (option a). From a careful comparison of ab initio and AM1 data for these molecules and smaller model systems,44 we expect that AM1 overestimates the C‚‚‚C distances slightly and that the AM1/MM3/A (option a) values are therefore probably too large by about 0.1 Å. The AM1 sequence of increasing C‚‚‚C distances (3 < 2 < 1 < 4 < 5) is, however, nicely reproduced by the QM/MM model. Even the differences between absolute values are very similar for AM1 and AM1/MM3/A (option a). Both methods further agree quantitatively in the nonbonded C‚‚‚H distances of 1-3. Compared to AM1, the QM/MM model yields slightly larger C‚‚‚H distances for 4 and 5. This reflects the larger discrepancies between the AM1 and AM1/ MM3/A relative activation energies calculated for these molecules (see Table 5). Both the QM/MM model and AM1 show consistent sequences of activation energies and C‚‚‚H distances. A correlation between these parameters has indeed been established,44 supporting the assumption that the MM cages merely act as geometrical anchors constraining the QM reactive center geometry. A closer examination of the QM/MM results, however, reveals a more complex situation (see Figure 5). While the QM energy differences ∆EQM(Y-L) for the reactive center follow the trend provided by the total activation energies ∆EAtot(X-Y), they account for only 40-50% quantitatively. The link atom corrections ∆ELINK contribute significantly, both by pronouncing the differences between the activation energies and by increasing their absolute values. This example again stresses the importance of ELINK as part of the total energy expression. The hydride transfer reactions of molecules 1-5 have also been studied by TSM (transition state modeling).101,102 This gives us the opportunity to comment on the relation between QM/MM and TSM approaches. Both attempt a quantum mechanical treatment of the reactive center, but in conceptually different ways. TSM applies two separately calibrated force fields for reactants and transition states, using QM geometries of model compounds as penalty functions. The QM/MM treatment, on the other hand, directly includes the QM model 10590 J. Phys. Chem., Vol. 100, No. 25, 1996 Bakowies and Thiel Figure 6. Substituted 7-norbornanones chosen for the QM/MM study. as integral part of the approach. Since the TSM studies and our AM1/MM3/A (option a) calculations are based on the same QM transition state model (CH2O + H- + CH2O) the results may be compared directly: They are apparently of similar overall quality, with a slight edge for the QM/MM results (see Table 5). While TSM is certainly more economical in its application, QM/MM approaches offer some conceptual advantages which make them more attractive for general purposes: First, no specific parametrization is required. Second, all stationary points are consistently treated in the same way. Since TSM uses two different force fields for reactants and transition states, it yields activation energies which are meaningless as absolute numbers. Only relative values may be compared. Third, QM/MM treats transition states in a physically correct way as maxima of the reaction coordinate, contrary to TSM which approximates them as minima of a constrained MM potential surface. Fourth, QM/MM studies are not restricted to specifically parametrized reaction channels. Finally, only QM/MM approaches allow one to model the polarization of the QM charge density due to MM point charges. 4.5 Nucleophilic Addition to Carbonyl Compounds. The nucleophilic addition of metal hydrides or metal alkyls to carbonyl compounds is similar in nature to the hydride transfer reaction discussed above, but it affords the intermediate formation of at least one dipole complex. The reaction mechanism has been studied theoretically in some detail,105-107 and much work has been devoted to the origin of stereocontrol (see, e.g., refs 108-110). We examined the QM/MM models B and C and tested their ability to reproduce substituent effects on mechanistic details such as activation energies, transition structures, stereochemical preferences, and the formation of specific dipole complexes. Here we discuss energetic aspects in some detail and give a short summary of the remaining results. A full report may be found elsewhere.44 MNDO,111 MNDO/MM3,111,112 and ab initio calculations,82 (RHF/6-31G* and MP2/6-31G*//RHF/6-31G* 1) have been carried out for the reactions of formaldehyde 6, acetone 7, and substituted 7-norbornanones 8-10 with monomeric lithium hydride113 (see Figure 6). In all QM/MM calculations for 7-10, the subunit (CH2O + LiH) is treated quantum mechanically while the remainder is described classically. The same bond topology has been used for any of the stationary points, employing carbonyl instead of alkoxy parameters even for the product. The lithium parameter for MNDO electrostatic potentials (RLi ) 1.11988 Å-1) has been obtained by the parametrization procedure described in ref 45, using LiH, LiCH3, and LiOH as reference molecules. All calculations refer to the C//B level if not stated otherwise. While molecules 6-9 consist of only one conformer each, five conformations need to be considered for molecule 10, resulting from different orientations of the two formyl groups. Unlike both MNDO and MM3, the hybrid MNDO/MM3 approach succeeds in reproducing their energetical sequence as predicted by ab initio methods, although the covered range of energies is underestimated (4.2 kcal/mol vs 6.1 and 6.8 kcal/ mol at the RHF/6-31G* and MP2/6-31G* levels, see supporting Figure 7. Nucleophilic addition of LiH to formaldehyde: Stationary points (RHF/6-31G* geometries). TABLE 7: Reaction Profile of the Nucleophilic Addition of LiH to 6-10a method MNDO MNDO/MM3/C//B RHF/6-31G* MP2/6-31G*//RHF/6-31G* molecule b 6 7 8 9 10 6b 7 8 9 10 6c 7d 8 9 10 6 7 8 9 10 RfD DfT TfP -18.0 -17.8 -16.1 -16.0 -14.5 -18.0 -21.4 -21.0 -21.1 -18.7 -19.8 -23.9 -24.1 -24.5 -22.1 -19.9 -24.3 -24.1 -24.6 -22.3 4.3 12.0 17.5 18.9 15.7 4.3 5.5 8.2 7.9 3.3 3.6 10.0 10.2 11.4 5.3 3.5 4.9 3.6 4.4 -1.4 -35.5 -32.8 -38.0 -38.7 -39.5 -35.5 -48.4 -52.3 -51.6 -51.5 -36.8 -29.7 -34.2 -34.6 -35.3 -43.5 -33.9 -38.5 -38.5 -38.4 a Difference in energy or heat of formation (kcal/mol) for the step indicated. T and P always refer to the syn oriented attack. b The QM/ MM and QM treatments are identical by definition. c See also ref 106. d See also: Wu, Y.-D.; Houk, K. N. J. Am. Chem. Soc. 1987, 109, 908. information for details). The energetically most favorable conformer shows nearly orthogonally oriented formyl groups and has no symmetry. The more favorable of the conformers with Cs symmetry has nearly ecliptic CO and CC bonds. It has been chosen for the present study since symmetry facilitates reasonably accurate ab initio reference calculations. The investigated reaction path has Cs symmetry throughout. All theoretical approaches applied show 4 distinct stationary points114 for any of the substrates 6-10: reactants R, dipole complexes D with almost linear COLiH units, four center transition states T, and products P (see Figure 7 and Table 7). The formation of the dipole complex D is generally quite exothermic. The RHF and MP2 levels of theory consistently predict an energy gain of 20-25 kcal/mol which is probably too large because of the basis set superposition error. The intrinsic activation energy (D f T) is very low (4-11 kal/ mol) at the RHF level and is further reduced by electron correlation. MP2 predicts even a negative barrier for the reaction of 10. There is, however, an additional dipole complex lower in energy (-2.1 kcal/mol) in this case which shows a bent COLiH unit resembling the transition state geometry (see supporting information). Large amounts of energy are released in the product formation step (T f P, RHF: 30-37 kcal/mol, MP2: 34-44 kcal/mol), resulting in a very exothermic overall reaction. The transition state is generally formed with a net gain of energy (R f T). Several differences in the reaction Quantum Mechanical and Molecular Mechanical Hybrid Models TABLE 8: Energy Contributions to the Reaction Profile: MNDO/MM3/C//Ba coul(X, E Eind(Y) Eind(X) ELINK a Y) molecule R D T P 7 8 9 10 7 8 9 10 7 8 9 10 7 8 9 10 -10.0 -4.6 -4.7 -2.2 -6.9 -2.7 -2.6 -0.3 -4.4 -3.5 -3.7 -1.2 -1.4 -1.2 -1.1 -0.8 -12.4 -6.4 -6.4 -2.0 -6.9 -2.7 -2.6 -0.3 -6.0 -5.3 -5.7 -2.5 -0.8 -0.7 -0.7 -0.5 -10.5 -4.1 -5.1 -4.1 -6.9 -2.6 -2.6 -0.3 -3.9 -3.4 -3.5 -1.6 -5.5 -4.8 -4.8 -4.5 -6.7 -2.2 -2.6 -1.9 -6.7 -2.6 -2.6 -0.2 -2.5 -1.7 -1.8 -0.3 -33.1 -32.8 -32.6 -31.5 All entries are in kcal/mol. T and P refer to the syn oriented attack. profiles of the substrates 6-10 are noticeable: (1) The complexation energy is lowest for formaldehyde 6, followed by the diformyl substituted 7-norbornanone 10. (2) 10 has an exceptionally low intrinsic activation energy, the same is observed for 6 at the RHF level of theory. (3) Acetone 7 shows the lowest gain of energy for the product formation step. MNDO/MM3 benefits from the satisfactory MNDO results for 6 and reproduces most of the trends observed in the ab initio calculations. The QM/MM model yields intrinsic activation energies which are in between the RHF and MP2 results, and predicts overall activation energies (referring to reactants rather than to dipole complexes) which are close to the RHF values. Most differences in the ab initio reaction profiles for 7-10 are attributed to electrostatic and induction effects by the QM/MM model (see Table 8). Compared to the unsubstituted 7-norbornanone 8, for example, the diformyl substituted molecule 10 shows less attractive Coulomb interactions for the reactant (2.4 kcal/mol) and for the dipole complex (4.4 kcal/mol). The induction energies Eind(X) + Eind(Y) are generally lower for 10 than for 8, with only small variations along the reaction path (4.5 ( 0.7 kcal/mol), resulting in both a lower complexation energy and a lower intrinsic barrier for 10. The least exothermic product formation step occuring for 7 may similarly be attributed to electrostatic effects (see Table 8). MNDO/MM3 consistently overestimates the stability of the products by 10-15 kcal/mol. This is completely due to the inappropriate representation of alkoxy groups by carbonyl bonded (X,Y).44 The parameters in the MM coupling term EMM large out of plane bending force constant associated with sp2 carbon atoms does not allow the required full pyramidalization of the alkoxy unit. Unlike the MM carbon atom, its substitute link atom experiences only a weak QM bending force and therefore adopts a position consistent with a tetrahedral sp3 geometry. The link atom correction ELINK for the QM geometry of Y-L is consequently too high and leads to excessive stabilization of the product. The same artifact also occurred in the QM/MM treatment of the hydride transfer reactions, but was less obvious since the errors for reactants and products were nearly compensated by smaller errors for two slightly bent carbonyl groups in the transition state. The problem discussed here should only appear for QM/MM divisions close to the reactive center, and even then it can easily be removed by appropriate parameter scaling. It seems worth mentioning that MNDO/MM3 performs better than MNDO in various aspects: For the first two steps (R f D, D f T), the QM/MM model yields energy differences in J. Phys. Chem., Vol. 100, No. 25, 1996 10591 much closer agreement with the ab initio values and correctly estimates the transition states to be lower in energy than the reactants. MNDO fails in three out of five cases. The QM/ MM model further predicts two additional dipole complexes, with the COLiH unit being bent either within the CsC(dO)sC plane (molecule 7) or perpendicular to it (molecule 10) (see supporting information). Both dipole complexes have been verified by RHF/6-31G* but cannot be located on the MNDO potential surface. Unlike MNDO, the QM/MM model qualitatively reproduces the relative energies of products resulting from syn and anti attack of LiH to 9 and 10 (see supporting information). It fails, however, to reproduce relative transition state energies and is thus unreliable in predicting the diastereoselectivities for this reaction (see supporting information). This is surprising since electrostatic effects have been used to explain syn or anti preferences,109 and the QM/MM approaches have so far been successful in modeling these effects. A careful examination of the ab initio and QM/MM results44 shows that the Coulomb forces between the QM and MM fragments are much less significant in directing the side of LiH attack than in a previous model44,109 where LiH is represented by classical point charges interacting with the ketone. The ability of our QM/MM models to reproduce trends in geometries has been thoroughly checked for this reaction. Selected results are given in the supporting information. Apart from a few exceptions, both the influence of MM substituents on the reactive center geometry and changes in the substituent geometries occurring along the reaction coordinate are reproduced qualitatively. The MNDO/MM3 geometries are not very accurate in an absolute sense, however, since they suffer from shortcomings in the MNDO parametrization115 of lithium.116 MNDO predicts LiH distances which are too short and LiO distances which are too long, and these errors are present in both the MNDO and the MNDO/MM3 results. 4.6 Nucleophilic Ring Cleavage of Oxiranes. The nucleophilic ring cleavage of unsymmetrically alkyl substituted oxiranes is known to proceed regiospecifically. While the attack at the less substituted carbon atom in basic media is commonly explained by steric effects directing the pathway of a SN2 reaction,117 the more facile cleavage at the substituted carbon atom in acidic media117,118 has caused a controversial discussion.117-125 Kinetic studies favored a SN1 mechanism120 in which the regioselectivity is caused by electronic stabilization (“+I effect”) of the intermediate carbenium ion. The observed Walden inversion,119 however, could only be rationalized with a SN2 mechanism, giving rise to the hypothesis of a “borderline SN2” pathway117 which has, however, repeatedly been criticized.119,122 The reaction of 2,2-dimethyloxirane with OH- (basic, reaction (a)) and of protonated 2,2-dimethyloxirane with H2O (acidic, reaction (b)) has been studied by MNDO/MM3/C//B to examine whether a simple QM/MM model can reproduce and explain the experimental observations. Only MNDO calculations have been carried out for reference, since we focus on qualitative rather than quantitative aspects. The study has further been restricted to the first part of the reaction, leading from reactants R to dipole complexes D and transition states T (see Figure 8). In the QM/MM treatment, the classical region comprises the two methyl groups. None of the CO bonds is considered in the MM part of the QM/MM coupling term to ensure a consistent and unified description for all reaction paths. The ring carbon atoms are represented by alkene parameters to account for their (partial) sp2 character.122 The MNDO and MNDO/MM3/C//B results are collected in Tables 9 and 10, respectively. Under base catalysis, the attack 10592 J. Phys. Chem., Vol. 100, No. 25, 1996 Bakowies and Thiel experimentally observed regioselectivities. It is interesting to note that the simpler QM/MM model A prefers attack at the less substituted carbon atom for both reactions a and b. It further yields much smaller differences in geometry for the two transition states resulting from acid-catalyzed attack. These results provide some insight in the origin of regioselectivity. While minimization of steric repulsion appears to be the driving force in basic media, Coulomb interactions between the QM and MM fragments prevail in the acid-catalyzed reaction. They are responsible for the large changes in geometry (supporting the “borderline SN2” hypothesis) and facilitate the attack at the sterically congested site. This explanation is closely related to the empirical argumentation discussed above, except that inductive (charge transfer) effects are replaced by electrostatic effects which, however, show the same trend for obvious reasons. Figure 8. Ring cleavage of oxiranes: Reactants R, dipole complexes D, and transition structures T; (a) base catalysis, (b) acid catalysis. The activation energies and activation enthalpies are defined as follows: ∆Eq1 ) E(T) - E(R), ∆Eq2 ) E(T) - E(D), ∆∆Hq1 ) ∆Hf0(T) - ∆Hf0(R), ∆∆Hq2 - ∆Hf0(T) - ∆Hf0(D). TABLE 9: Nucleophilic Ring Cleavage of Oxiranes: MNDO Resultsa reaction R attackb ∆∆Hq1 ∆∆Hq2 rform rbreak a a a bc b b H, H CH3, CH3 CH3, CH3 H, H CH3, CH3 CH3, CH3 1 1 2 1 1 2 11.8 25.5 12.6 6.2 -1.5 12.9 17.4 31.2 18.3 12.5 4.0 18.4 1.942 2.000 1.897 3.021 3.606 3.077 1.571 1.674 1.533 1.943 1.798 2.034 a Activation enthalpies ∆∆Hq in kcal/mol, transition structure parameters in Å. See Figure 8 for reference. b See Figure 8 for reference. c See also ref 123. TABLE 10: Nucleophilic Ring Cleavage of Oxiranes: MNDO/MM3/C//B Resultsa reaction R attackb ∆Eq1 ∆Eq2 rform rbreak a a b b CH3, CH3 CH3, CH3 CH3, CH3 CH3, CH3 1 2 1 2 21.8 13.7 -2.3 4.2 31.5 23.3 3.3 9.8 1.980 1.884 3.226 2.982 1.684 1.585 1.884 2.041 a MNDO/MM3(CH , CH ). Activation energies ∆Eq in kcal/mol, 3 3 transition structure parameters in Å. See Figure 8 for reference. b See Figure 8 for reference. at the unsubstituted carbon atom is strongly preferred (by 12.9 kcal/mol (MNDO) and 8.1 kcal/mol (MNDO/MM3), respectively). Both methods also agree that acid catalysis favors attack at the substituted carbon atom (by 14.4 and 6.5 kcal/mol, respectively). As expected, the lower activation energy corresponds to an earlier transition state as far as the breaking bond is concerned. The forming bond, on the other hand, is always longer for the attack at the substituted carbon atom, reflecting the considerable steric repulsion between nucleophile and methyl groups. The QM/MM model reproduces the QM trends nicely, but yields less pronounced differences, both in geometry and in energy, for the alternative pathways. The geometries may not be very accurate in either case, since MNDO is known to overestimate nonbonded distances for the acid-catalyzed reaction of unsubstituted oxirane44,123 while it underestimates them for the base-catalyzed reaction.44,126 The considerably smaller MNDO/MM3 values for reaction (a) are certainly more realistic but probably still too large. Nevertheless, the MNDO/MM3/C//B results may be regarded as satisfactory since they are in qualitative agreement with the 5. Discussion and Conclusions We have presented a hierarchy of three models to combine quantum mechanical (QM) and molecular mechanical (MM) approaches. They are designed to reduce the QM treatment of large molecules to the electronically important fragment which then interacts in a perturbative fashion with the molecular mechanically described remainder. While model A represents a simple mechanical embedding of the QM fragment, more refined treatments of electrostatic and induction interactions are included in models B and C. In contrast to earlier QM/MM approaches, the proposed models include an explicit correction for interactions involving the link atom which substitutes the MM fragment in the QM treatment. This correction appears to be an important part of the total energy expression, and several test cases have shown that it can improve the results considerably (see, e.g., sections 4.3 and 4.4). The link atom correction behaves reasonably well for geometries close to the optimized QM structures while it is inappropriate for geometries far off the minimum. This prevents the use of the total energy expression for geometry optimizations and makes the separate definition of a potential energy inevitable. Inadequate MM parameters for atoms at the QM/MM boundary may also lead to ill-behaved link atom corrections. This causes problems whenever the QM/MM treatment of a chemical reaction requires a QM/MM boundary close to the reactive center with MM parameters which are kept fixed for the entire reaction path (see, e.g., section 4.5). Hence, future applications should consider an appropriate scaling of the MM parameters in question. The angle between the bond connecting the QM and MM fragments and the bond of the frontier QM atom with the link atom provides a useful check for the quality of these MM parameters. It should not be larger than a few degrees, indicating a wellbalanced MM description of the boundary region.44 QM/MM approaches with an explicit link atom correction have the formal disadvantage that they cannot be used in MD simulations which require consistent total energies and gradients. This problem may be circumvented by deliberately shifting the QM/MM boundary as far away from the reactive center as is necessary to achieve a sufficiently constant link atom correction for the entire reaction path. In this case, it seems justified to replace the total energy by the potential energy which leads to consistent energies and gradients. The effects of this approximation need to be checked in any given application. Model B includes a quantum chemical treatment of Coulomb interactions between the QM and MM fragments. As a refinement of earlier QM/MM approaches, we have adopted parametrized models to calculate QM electrostatic potentials and MM partial charges which have both been calibrated against Quantum Mechanical and Molecular Mechanical Hybrid Models RHF/6-31G* reference data.45 In the majority of cases, model B successfully reproduces (ab initio or experimental) trends arising from electrostatic effects which indicates a reasonable description of Coulomb interactions. There are, however, some deficiencies of model B: Electrostatic interactions involving strongly charged MM atoms close to the QM/MM boundary tend to be overestimated and to induce charge flux between the QM fragment and the link atom which generally lowers the QM/MM total energy.44 The link atom approach does not provide any possibility to prevent this unphysical side effect. Future developments should attempt to ameliorate this problem. This might already be achieved by introducing secondary conditions in the charge equilibration model which reduce the MM charges at the QM/MM boundary. Such a refinement would empirically account for the physically well-known damping behavior of short-range interactions and simultaneously reduce the undesirable charge flux. In the present implementation, the calculation of relative energies benefits from error cancellation.44 Problems connected with either inappropriate MM parameters for the QM/MM boundary or the poor representation of short range interactions are likely to cause errors of similar magnitude for closely related conformations or stationary points. The successful application of the proposed QM/MM models to various problems of chemical structure and reactivity has indeed shown that the more interesting relative energies (or relative geometries) are barely influenced by these shortcomings. As a general guideline, it is obviously advantageous to put the QM/MM boundary as far away from the reactive center as is computationally affordable in order to minimize such errors and to allow for an optimum error cancelation. Model C includes the polarization of the MM fragment. It adopts the physically justifiable treatment of Thole63 to describe induced dipole interactions which is considered as a significant improvement over the older treatment proposed by Applequist.62 Although some refinement is still possible for the description of short range polarization at the QM/MM boundary, model C currently provides one of the most advanced treatments of polarization in semiempirical QM/MM approaches. The consideration of MM polarization appears to be crucial in applications involving charged QM fragments which generate large electric fields. This has been exemplified for the calculation of deprotonation enthalpies where considerable qualitative improvements have been found compared with other QM/MM treatments. The proposed QM/MM approaches are generally not accurate in a quantitative sense, due to deficiencies in the interaction models and in the underlying QM and MM methods. In most cases, however, they provide sufficiently reliable qualitative results so that they can be used for an interpretation of experimental data, especially with regard to trends in related molecules. Acknowledgment. This work has been supported in part through the Alfried Krupp Förderpreis. Supporting Information Available: Additional data are provided for the protonation energies and deprotonation enthalpies of alcohols (Tables S1-S3) and for the nucleophilic addition reaction of LiH with ketones (Tables S4-S9) (11 pages). Ordering information is given on any current masthead page. References and Notes (1) Hehre, W. J.; Radom, L.; Schleyer, P. v. R.; Pople, J. A. Ab Initio Molecular Orbital Theory; Wiley & Sons, Inc.: New York, 1986. J. Phys. Chem., Vol. 100, No. 25, 1996 10593 (2) Ziegler, T. Chem. ReV. 1991, 91, 651. (3) Thiel, W. AdV. Chem. Phys. 1996, 93, 703. (4) Bakowies, D.; Thiel, W. J. Am. Chem. Soc. 1991, 113, 3704. (5) Bakowies, D.; Bühl, M.; Thiel, W. J. Am. Chem. Soc. 1995, 117, 10113. (6) Burkert, U.; Allinger, N. L. Molecular Mechanics; American Chemical Society: Washington, D.C., 1982. (7) Warshel, A.; Levitt, M. J. Mol. Biol. 1976, 103, 227. (8) Field, M. J.; Bash, P. A.; Karplus, M. J. Comput. Chem. 1990, 11, 700. (9) Vasilyev, V. V.; Bliznyuk, A. A.; Voityuk, A. A. Int. J. Quantum Chem. 1992, 44, 897. (10) Théry, V.; Rinaldi, D.; Rivail, J.-L.; Maigret, B.; Ferenczy, G. J. Comput. Chem. 1994, 15, 269. (11) Thompson, M. A.; Glendening, E. D.; Feller, D. J. Phys. Chem. 1994, 98, 10465. (12) Thompson, M. A.; Schenter, G. K. J. Phys. Chem. 1995, 99, 6374. (13) Stanton, R. V.; Hartsough, D. S.; Merz, K. M., Jr. J. Comput. Chem. 1995, 16, 113. (14) Bernardi, F.; Olivucci, M.; Robb, M. A. J. Am. Chem. Soc. 1992, 114, 1606. (15) Åqvist, J.; Warshel, A. Chem. ReV. 1993, 93, 2523. (16) Singh, U. C.; Kollman, P. A. J. Comput. Chem. 1986, 7, 718. (17) Bash, P. A.; Field, M. J.; Karplus, M. J. Am. Chem. Soc. 1987, 109, 8092. (18) Gao, J. J. Phys. Chem. 1992, 96, 537, 6432. (19) Gao, J.; Xia, X. Science 1992, 258, 631. (20) Gao, J.; Pavelites, J. J. J. Am. Chem. Soc. 1992, 114, 1912. (21) Gao, J. Int. J. Quantum Chem.: Quantum Chem. Symp. 1993, 27, 491. (22) Gao, J.; Luque, F. J.; Orozco, M. J. Chem. Phys. 1993, 98, 2975. (23) Gao, J. J. Am. Chem. Soc. 1993, 115, 2930. (24) Gao, J.; Xia, X. J. Am. Chem. Soc. 1993, 115, 9667. (25) Stanton, R. V.; Hartsough, D. S.; Merz, K. M., Jr. J. Phys. Chem. 1993, 97, 11868. (26) Gao, J. J. Am. Chem. Soc. 1994, 116, 1563, 9324. (27) Liu, H.; Shi, Y. J. Comput. Chem. 1994, 15, 1311. (28) Liu, H.; Müller-Plathe, F.; van Gunsteren, W. F. J. Chem. Phys. 1995, 102, 1722. (29) Stanton, R. V.; Little, L. R.; Merz, K. M., Jr. J. Phys. Chem. 1995, 99, 483. (30) Hartsough, D. S.; Merz Jr., K. M. J. Phys. Chem. 1995, 99, 384. (31) Thompson, M. A. J. Phys. Chem. 1995, 99, 4794. (32) Bash, P. A.; Field, M. J.; Davenport, R. C.; Petsko, G. A.; Ringe, D.; Karplus, M. Biochemistry 1991, 30, 5826. (33) Waszkowycz, B.; Hillier, I. H.; Gensmantel, N.; Payling, D. W. J. Chem. Soc., Perkin Trans. 2 1991, 225, 1819, 2025. (34) Vasilyev, V. V. J. Mol. Struct. (THEOCHEM) 1994, 304, 129. (35) Elcock, A. H.; Lyne, P. D.; Mulholland, A. J.; Nandra, A.; Richards, W. G. J. Am. Chem. Soc. 1995, 117, 4706. (36) Hartsough, D. S.; Merz, Jr. K. M. J. Phys. Chem. 1995, 99, 11266. (37) Dewar, M. J. S.; Thiel, W. J. Am. Chem. Soc. 1977, 99, 4899. (38) Dewar, M. J. S.; Thiel, W. J. Am. Chem. Soc. 1977, 99, 4907. (39) Dewar, M. J. S.; Zoebisch, E. G.; Healy, E. F.; Stewart, J. J. P. J. Am. Chem. Soc. 1985, 107, 3902. (40) Allinger, N. L.; Yuh, Y. H.; Lii, J.-H. J. Am. Chem. Soc. 1989, 111, 8551. (41) Lii, J.-H.; Allinger, N. L. J. Am. Chem. Soc. 1989, 111, 8566, 8576. (42) Thiel, W. Tetrahedron 1988, 44, 7393. (43) See, e.g.: Allinger, N. L.; Fan Y. J. Comput. Chem. 1993, 14, 655 and references cited therein. (44) Bakowies, D. Hybridmodelle zur Kopplung quantenchemischer und molekülmechanischer Verfahren; Ph.D. Thesis, Zürich, Hartung-Gorre Verlag: Konstanz, 1994. (45) Bakowies, D.; Thiel, W. J. Comput. Chem. 1996, 17, 87. (46) Rappé, A. K.; Casewit, C. J.; Colwell, K. S.; Goddard, W. A., III; Skiff, W. M. J. Am. Chem. Soc. 1992, 114, 10024. (47) Weiner, P. K.; Kollman, P. A. J. Comput. Chem. 1981, 2, 287. (48) Weiner, S. J.; Kollman, P. A.; Case, D. A.; Singh, U. C.; Ghio, C.; Alagano, G.; Profeta, S., Jr.; Weiner, P. J. Am. Chem. Soc. 1984, 106, 765. (49) Weiner, S. J.; Kollman, P. A.; Nguyen, D. T.; Case, D. A. J. Comput. Chem. 1986, 7, 230. (50) Cornell, W. D.; Cieplak, P.; Bayly, C. I.; Gould, I. R.; Merz, K. M., Jr.; Ferguson, D. M.; Spellmeyer, D. C.; Fox, T.; Caldwell, J. W.; Kollman, P. A. J. Am. Chem. Soc. 1995, 117, 5179. (51) Brooks, B. R.; Bruccoleri, R. E.; Olafson, B. D.; States, D. J.; Swaminathan, S.; Karplus, M. J. Comput. Chem. 1983, 4, 187. (52) Allinger, N. L.; Tai, J. C. J. Am. Chem. Soc. 1965, 87, 2081. (53) Allinger, N. L.; Tai, J. C.; Stuart, T. W. Theor. Chim. Acta 1967, 8, 101. (54) Sprague, J. T.; Tai, J. C.; Yuh, Y.; Allinger, N. L. J. Comput. Chem. 1987, 8, 581. (55) Allinger, N. L.; Li, F.; Yan, L.; Tai, J. C. J. Comput. Chem. 1990, 11, 868. 10594 J. Phys. Chem., Vol. 100, No. 25, 1996 (56) Dosen-Micovic, L.; Allinger, N. L. J. Am. Chem. Soc. 1983, 105, 1723. (57) Rychnovsky, S. D.; Yang, G.; Powers, J. P. J. Org. Chem. 1993, 58, 5251. (58) Welti, M. Ph.D. Thesis, ETH Zürich, 1987. (59) Williams, D. E. ReV. Comp. Chem. 1991, 2, 219. (60) Dang, L. X.; Rice, J. E.; Caldwell, J.; Kollman, P. A. J. Am. Chem. Soc. 1991, 113, 2481. (61) Silberstein, L. S. Philos. Mag. 1917, 33, 92, 215, 521. (62) Applequist, J.; Carl, J. R.; Fung, K.-K. J. Am. Chem. Soc. 1972, 94, 2952. (63) Thole, B. T. Chem. Phys. 1981, 59, 341. (64) Stone, A. J. Chem. Phys. Lett. 1989, 155, 102. (65) Buckingham, A. D. In Intermolecular Interactions: From Diatomics to Biopolymers; B. Pullmann, Ed.; J. Wiley & Sons: Chichester, 1978; pp 1-67. (66) Tapia, O.; Goscinski, O. Mol. Phys. 1975, 29, 1653. (67) Thole, B. T.; van Duijnen, P. T. Theor. Chim. Acta 1980, 55, 307. (68) De Vries, A. H.; Van Duijnen, P. T.; Juffer, A. H.; Rullmann, J. A. C.; Dijkman, J. P.; Merenga, H.; Thole, B. T. J. Comput. Chem. 1995, 16, 37. (69) Tapia, O. J. Mol. Struct. (THEOCHEM) 1991, 226, 59. (70) van Duijnen, P. T.; Rullmann, J. A. C. Int. J. Quantum Chem. 1990, 38, 181. (71) Thiel, W.: Program MNDO91; Universität Wuppertal, 1991. (72) Allinger, N. L.; Yuh, Y. H.; Lii, R.: Program MM3(89); Technical Utilization Corporation, Powell, OH 43065, 1989. (73) Giessner-Prettre, C.; Pullman, A. Theor. Chim. Acta 1972, 25, 83. (74) Luque, F. J.; Orozco, M. Chem. Phys. Lett. 1990, 168, 269. (75) Luque, F. J.; Illas, F.; Orozco, M. J. Comput. Chem. 1990, 11, 416. (76) Alhambra, C.; Luque, F. J.; Orozco, M. J. Comput. Chem. 1994, 15, 12. (77) Besler, B. H.; Merz, K. M.; Kollman, P. A. J. Comput. Chem. 1990, 11, 431. (78) Orozco, M.; Luque, F. J. J. Comput. Chem. 1990, 11, 909. (79) Ford, G. P.; Wang, B. J. Comput. Chem. 1993, 14, 1101. (80) Rappé, A. K.; Goddard, W. A., III. J. Phys. Chem. 1991, 95, 3358. (81) Pople, J. A.; Krishnan, R.; Schlegel, H. B.; Binkley, J. S. Int. J. Quantum Chem.: Quantum Chem. Symp. 1979, 13, 225. (82) All ab initio calculations have been carried out using the Gaussian92 suite of programs: Frisch, M. J.; Trucks, G. W.; Head-Gordon, M.; Gill, P. M. W.; Wong, M. W.; Foresman, J. B.; Johnson, B. G.; Schlegel, H. B.; Robb, M. A.; Replogle, E. S.; Gomperts, R.; Andres, J. L.; Raghavachari, K.; Binkley, J. S.; Gonzalez, C.; Martin, R. L.; Fox, D. J.; Defrees, D. J.; Baker, J.; Stewart, J. J. P.; Pople, J. A.: Program Gaussian92, Revision B; Gaussian92, Inc., Pittsburgh PA, 1992. (83) Olivella, S.; Urpi, F.; Vilarassa, J. J. Comput. Chem. 1984, 5, 230. (84) Dewar, M. J. S.; Dieter, K. M. J. Am. Chem. Soc. 1986, 108, 8075. (85) Lias, S. G.; Bartmess, J. E.; Liebman, J. F.; Holmes, J. L.; Levin, R. D.; Mallard, W. G. J. Phys. Chem. Ref. Data 1988, 17S1, 1. (86) Munson, M. S. B. J. Am. Chem. Soc. 1965, 87, 2332. (87) Brauman, J. I.; Blair, L. K. J. Am. Chem. Soc. 1968, 90, 6561. (88) Brauman, J. I.; Blair, L. K. J. Am. Chem. Soc. 1970, 92, 5986. (89) Taft, R. W. Prog. Phys. Org. Chem. 1983, 14, 247. (90) Smith, S. F.; Chandrasekhar, J.; Jorgensen, W. L. J. Phys. Chem. 1982, 86, 3308. (91) The term “inductive effect” relies on empirical concepts frequently used in organic chemistry, but is generally not precisely defined. It describes some sort of intramolecular charge transfer and should thus never be mixed up with induction which has a physically exact definition and is synonymous with polarization. (92) Hine, J.; Hine, M. J. Am. Chem. Soc. 1952, 74, 5266. Bakowies and Thiel (93) Bartmess, J. E.; Scott, J. A.; McIver, R. T., Jr. J. Am. Chem. Soc. 1979, 101, 6056. (94) Chauvel, J. P., Jr.; True, N. S. Chem. Phys. 1985, 95, 435. (95) Henry, R. S.; Riddell, F. G.; Parker, W.; Watt, C. I. F. J. Chem. Soc., Perkin Trans. 2 1976, 1549. (96) Craze, G.-A.; Watt, I. J. Chem. Soc., Perkin Trans. 2 1981, 175. (97) Cernik, R.; Craze, G.-A.; Mills, O. S.; Watt, I. J. Chem. Soc., Perkin Trans. 2 1982, 361. (98) Test calculations for some of the examples have indeed indicated negligible differences between the results obtained with models A-C. See chapter 8.2 in ref 44. (99) Parameters for open chains have also been used for four-membered rings whenever special ring parameters were not available in MM3(89). (100) Eksterowicz, J. E.; Houk, K. N. Chem. ReV. 1993, 93, 2439. (101) Sherrod, M. J.; Menger, F. M. J. Am. Chem. Soc. 1989, 111, 2611. (102) Eurenius, K. P.; Houk, K. N. J. Am. Chem. Soc. 1994, 116, 9943. (103) Cernik, R. V.; Craze, G.-A.; Mills, O. S.; Watt, I.; Whittleton, S. N. J. Chem. Soc., Perkin Trans. 2 1984, 685. (104) (a) Craze, G.-A.; Watt, I. Tetrahedron Lett. 1982, 9, 975. (b) Watt, I.; Whittleton, S. N.; Whitworth, S. M. Tetrahedron 1986, 42, 1047. (105) Bonaccorsi, R.; Palla, P.; Tomasi, J. J. Mol. Struct. (THEOCHEM) 1982, 87, 181. (106) Kaufmann, E.; Schleyer, P. v. R.; Houk, K. N.; Wu, Y.-D. J. Am. Chem. Soc. 1985, 107, 5560. (107) Nakamura, M.; Nakamura, E.; Koga, N.; Morokuma, K. J. Am. Chem. Soc. 1993, 115, 11016. (108) Wu, Y.-D.; Tucker, J. A.; Houk, K. N. J. Am. Chem. Soc. 1991, 113, 5018. (109) Paddon-Row, M. N.; Wu, Y.-D.; Houk, K. N. J. Am. Chem. Soc. 1992, 114, 10638. (110) Wong, S. S.; Paddon-Row, M. N. Aust. J. Chem. 1991, 44, 765. (111) Lithium parameters are available for MNDO,115 but not for AM1. (112) Van der Waals parameters for lithium have been taken from ref 46 and converted to the slightly different form of the Buckingham potential used in MM3:40 (Li) ) 0.0222 kcal/mol, rν(Li) ) 1.2255 Å. (113) Although the reaction in reality involves oligomeric forms of LiH, this does apparently not affect qualitative features of the reaction profile (Kaufmann, E. Ph.D. Thesis, Erlangen, 1989). Hence we may assume monomeric LiH in our calculations. (114) The movement of LiH in the dipole complex, the coupled rotations of the formyl groups in 10 or methyl groups in 9, and the rotation of the QM subunit CH2O with respect to the CO axis (QM/MM calculations) are characterized by shallow potentials and sometimes refer to imaginary frequencies. Since we are not interested in these subtle effects, we use the term transition state for any stationary point whose reaction coordinate corresponds to a negative eigenvalue of the hessian. (115) Thiel, W.: Program MNDOC; Quantum Chemistry Program Exchange: Bloomington, Indiana, 1982; QCPE Bull. 1982, 2, 63. (116) Anders, E.; Koch, R.; Freunscht, P. J. Comput. Chem. 1993, 14, 1301. (117) Parker, R. E.; Isaacs, N. S. Chem. ReV. 1959, 59, 737. (118) Long, F. A.; Pritchard, J. G. J. Am. Chem. Soc. 1956, 78, 2663. (119) Wohl, R. A. Chimia 1974, 28, 1. (120) Pritchard, J. G.; Long, F. A. J. Am. Chem. Soc. 1956, 78, 2667. (121) Pritchard, J. G.; Siddiqui, I. A. J. Chem. Soc., Perkin Trans. 2 1973, 452. (122) Dewar, M. J. S.; Ford, G. P. J. Am. Chem. Soc. 1979, 101, 783. (123) Ford, G. P.; Smith, C. T. J. Am. Chem. Soc. 1987, 109, 1325. (124) Ford, G. P.; Smith, C. T. J. Comput. Chem. 1989, 10, 568. (125) Ford, G. P.; Smith, C. T. J. Chem. Soc., Chem. Comm. 1987, 44. (126) Gronert, S. Int. J. Mass Spectrom. Ion Processes 1992, 117, 115. JP9536514