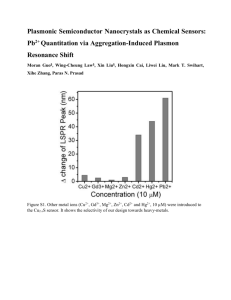
Analytical Small Zone Size Exclusion Chromatography some brief
advertisement

Analytical Small Zone Size Exclusion Chromatography ... some brief notes.
version 1.01 Last Edit RLK, 23 Dec 2009.
1. Select the size exclusion media appropriate to your problem.
2. Run your protein over the size exclusion column. Note that behavior of your protein may be
concentration dependent, so you may wish to vary the concentration. After this inject appropriate
size standards. Both Sigma and GE Healthcare sell protein standards for size exclusion
chromatography in lyophilized form. These proteins can be run together in suitable combinations to
save time if required. You also need to inject Blue Dextran 2000 (by itself) and a small molecule such
as Tyrosine (again by itself) onto the column.
3. Determine the elution volumes (Ve) of all species as the centre of the corresponding peak. The Void
volume of the column (V0) is the elution volume of Blue Dextran 2000. Knowing this, the internal
volume of the column (Vi) can be calculated from the eluted volume of Tyrosine, or similar small
molecule, which is V0 + Vi.
4. Calculate the partition coefficient (σ) for each species as follows:
v=
(1)
^ Ve - Vo h
^ Vih
Note that
σ = 0 for a molecule that’s totally excluded from the gel phase (e.g. Blue Dextran).
σ = 1 for a molecule that diffuses freely through the gel network with no restrictions (e.g. Tyrosine).
0 < σ < 1 for a molecule of intermediate size that doesn’t interact with the gel matrix
5. For the standards of known size, plot the cube root of the distribution coefficient (σ1/3) versus
hydrodynamic radius (RH) . For standard linear least squares (straight line fitting) it’s assumed by
most programs that the abscissa (x coordinate) is largely error-free, while the ordinate ( y
coordinate) contains the significant experimental error. In the situation here, it’s the distribution
coefficient that’s known most reliably, while the exact Hydrodynamic radius of the standards is more
uncertain (see the table below). So plot the cube root of the distribution coefficient (σ1/3) along
the x-axis.
Table 1. Size of protein SEC standards sold by GE Healthcare and Sigma.
Standard
~ Molecular Weight
Hydrodynamic radius
(According to primary sources)
Cytochrome C
(Equine Heart)
12400
18.7 {Larew, 1987, p15915}
18.5 {Walters, 1984, p15917}
18.0 {Clark, 2002, p15940}
18.2 {Bor Fuh, 1993, p15916}
Ribonuclease A
(Bovine pancreas)
13700
20.0 {Creeth, 1958, p15896}
19.0 {Nöppert, 1996, p15897}
17.0 {Kawahara, 1969, p15900}
17.6 {van Holde, 1958, p15914}
18.0 {Larew, 1987, p15915}
18.1 {Bor Fuh, 1993, p15916}
17.7 {Bor Fuh, 1993, p15916
18.4 {Walters, 1984, p15917}
Standard
~ Molecular Weight
Hydrodynamic radius
(According to primary sources)
Carbonic Anhydrase B
(Bovine erythrocytes)
29000
24.1 {Armstrong, 1966, p15899}
24.9 {Quinn, 1986, p15906}
Ovalbumin
(Hen egg)
44000
28.0 {Nemoto, 1993, p07762}
27.1 {Gibbs, 1991, p15908}
27.7 {Liu, 1993, p15910}
27.5 {Larew, 1987, p15915}
27.8 {Lamm, 1936, p15909}
30.0 {Walters, 1984, p15917}
Albumin
(Bovine Serum)
66000
34.5 {Bor Fuh, 1993, p15916}
35.9 {Bor Fuh, 1993, p15916}
34.0 {Walters, 1984, p15917}
35.7 {Raj, 1974, p15943}
Conalbumin
(Hen egg white)
75000
37.5 {Bezkorovainy, 1968, p15905}
Alcohol deydrogenase
(Yeast)
150000
42.2 {Bühner, 1969, p15944}
45.6 {HAYES, 1954, p15945}
Aldolase
(Rabbit muscle)
158000
49.8 {Kawahara, 1969, p15900}
Beta-Amylase
(Sweet Potato)
200000
48.2 {Liu, 1993, p15910}
48.9 {Liu, 1993, p15910}
Ferritin/ApoFerritin
(Horse spleen)
440000
58.1 {Liu, 1993, p15910}
62.2 {Liu, 1993, p15910}
60.5 {Clough, 1981, p15903}
53.7 {Walters, 1984, p15917}
65.9 {Walters, 1984, p15917}
66.1 {de Haën, 1987, p15947}
67.1 {de Haën, 1987, p15947}
Thyroglobulin
669000
86.1 {EDELHOCH, 1960, p15901}
83.3 {Liu, 1993, p15910}
81.4 {Liu, 1993, p15910}
92.3 {Liu, 1993, p15910}
(Bovine thyroid)
5. Fit a straight line to the data (see Winzor, 2003; Sigel and Monty, 1966), and determine the
hydrodynamic radius of the unknown protein using the linear fit.
References
Winzor. Analytical exclusion chromatography. J Biochem Biophys Methods (2003) vol. 56 (1-3) pp.
15-52
Siegel and Monty. Determination of molecular weights and frictional ratios of proteins in impure
systems by use of gel filtration and density gradient centrifugation. Application to crude preparations
of sulfite and hydroxylamine reductases. Biochim Biophys Acta (1966) vol. 112 (2) pp. 346-62