Activity: Recreating the Tree of Life Using Bioinformatics
advertisement
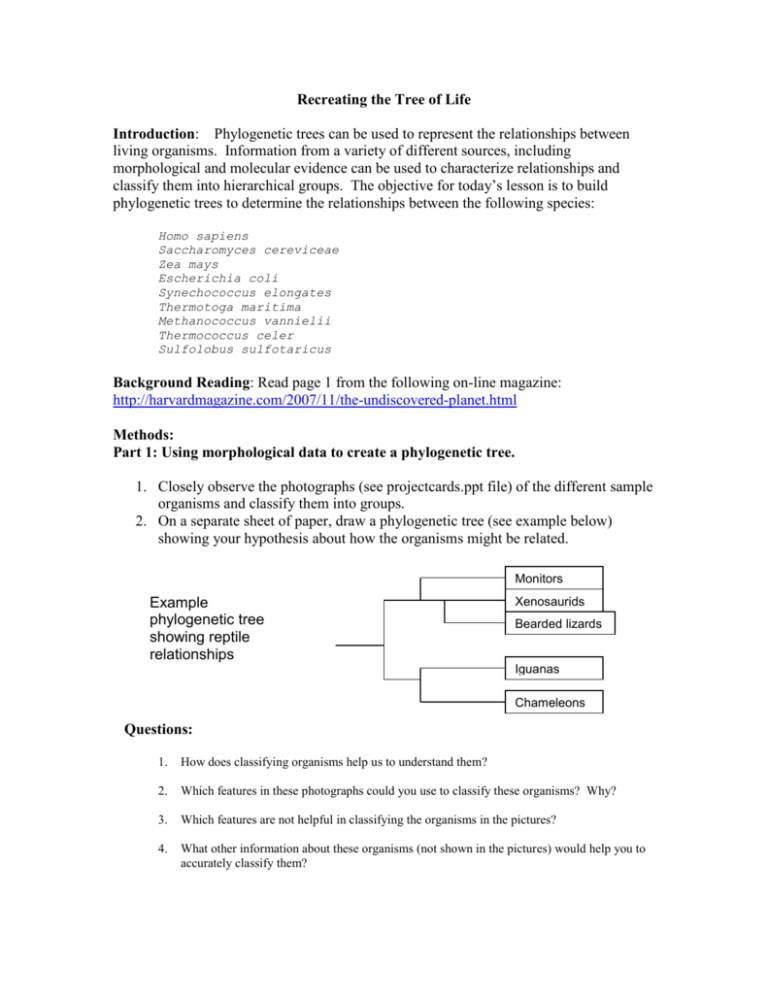
Recreating the Tree of Life Introduction: Phylogenetic trees can be used to represent the relationships between living organisms. Information from a variety of different sources, including morphological and molecular evidence can be used to characterize relationships and classify them into hierarchical groups. The objective for today’s lesson is to build phylogenetic trees to determine the relationships between the following species: Homo sapiens Saccharomyces cereviceae Zea mays Escherichia coli Synechococcus elongates Thermotoga maritima Methanococcus vannielii Thermococcus celer Sulfolobus sulfotaricus Background Reading: Read page 1 from the following on-line magazine: http://harvardmagazine.com/2007/11/the-undiscovered-planet.html Methods: Part 1: Using morphological data to create a phylogenetic tree. 1. Closely observe the photographs (see projectcards.ppt file) of the different sample organisms and classify them into groups. 2. On a separate sheet of paper, draw a phylogenetic tree (see example below) showing your hypothesis about how the organisms might be related. Monitors Example phylogenetic tree showing reptile relationships Xenosaurids Bearded lizards Iguanas Chameleons Questions: 1. How does classifying organisms help us to understand them? 2. Which features in these photographs could you use to classify these organisms? Why? 3. Which features are not helpful in classifying the organisms in the pictures? 4. What other information about these organisms (not shown in the pictures) would help you to accurately classify them? Part 2: Using a variety of sources to create a phylogenetic tree. 1. Look at the information about each organism (found on the back of each photograph). 2. Decide whether this new information SUPPORTS or REJECTS your original phylogenetic tree. 3. If your group decides that the information rejects or conflicts with your initial hypothesis, you should create a revised drawing of your tree. Questions: 1. What information did you use to classify? Explain why. 2. How might using morphological characteristics vs. a variety of sources produce different results? Part 3. Using molecular data to create a phylogenetic tree Instructions For Creating a Phylogenetic Tree based on 16s rRNA sequences. Our third phylogenetic tree will be prepared using a collection of bioinformatics tools available through the Biology Workbench at http://workbench.sdsc.edu/ . Your first step is to set up an account, by clicking on set up an account and following the directions. Once you have set up an account, follow the steps below: Step 1: Start a session 1. Scroll down until you see a series of boxes. 2. Click on the box labeled Session Tools. Now look through the menu of the long rectangular box and select Start New Session. Hit the Run button. Name the session “Tree of life”. 3. Select the box labeled Nucleotide Tools. Step 2: You are now ready to upload the rRNA sequences in the file: tree of life.txt 4. From the menu, select Add a New Nucleotide Sequence. A new box will appear on your screen. 5. Select Browse, then find the file named tree of life.txt and open it. 6. Select the Upload button. The sequences will be uploaded into your session window. 7. Go to the top or bottom of the screen and click on the Save button. You should now see a list of your sequences. Step 3: Creating a multiple alignment with ClustalW. 8. For each sequence, click in the box to the left of the sequence name to select it. Select each of the sequences in this manner. 9. From the menu box, select ClustalW. Select run. When the new screen appears, click on submit from the box on the screen. When the program has finished running, scroll down to view your alignment. Step 4: Creating a phylogenetic tree. 10. Find the window labeled Import Alignments and click on it. 11. Under the rectangle labeled Alignment Tools, select DrawGram. 12. Click on the small box nest to the list of aligned sequences and select submit. 13. Scroll down to see your phylogenetic tree. The tree can be dragged to a word processing file. Step 5: Support or Reject your hypothesis. 14. Decide whether information from rRNA sequence data SUPPORTS or REJECTS your original phylogenetic tree. 15. If your group decides that the information rejects or conflicts with your initial hypothesis, you should create a revised drawing of your tree. Questions: 1. How might using morphological characteristics vs. DNA sequences produce different results? 2. What are the advantages to using the morphological features such as those shown in the pictures or physical/ecological information in order to classify organisms? 3. What are some possible disadvantages to using morphological features or physical/ecological information in order to classify organisms? 4. What are the advantages to using DNA (or RNA) sequences in order to classify organisms? 5. What are some possible disadvantages to using DNA (or RNA) sequences in order to classify organisms? 6. Using DNA sequences to classify organisms is changing many of the traditional organization schemes (i.e. 3 domains instead of 5 kingdoms), is there still room for using traditional methods in 21st century Biology? 7. It would seem to be advantageous to compare the total DNA sequence of one organism to the total DNA sequence of another in order to most accurately compare and classify them. What problems/limitations can occur that prevent people from using such a comprehensive approach? 8. Explain the statement “rRNA is ‘just right’ for creating a universal tree of life”. 9. The casein gene codes for a protein that is found in milk. The sequence of this protein has been used to study mammalian phylogeny. Do you think that casein would be suitable for creating a universal tree of life? Why or why not? 10. Based on your final phylogenetic tree what conclusion can you draw about the relationships between the Eukarya, Archaea, and Bacteria?







