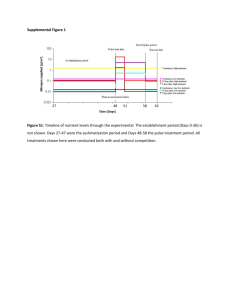
Supplementary Appendix 4 (doc 44K)

Appendix 4. Effects of nutrient addition and dispersal treatments on the community composition of prokaryotes and viruses.
Results of distance-based redundancy analyses relating Jaccard dissimilarity matrices calculated from presence-absence data of bacteria (T-RLP) and viruses
(RAPD) with the experimental treatments. Our inoculation design required us to do two analyses for each organism group: one analysis comparing NUT
H
- with the NUT
L
-mesocosms that had received the same inoculum, and one analysis comparing NUT
N
-containers with the corresponding NUT
L
-containers (see
Fig. 3.1 of Appendix 3 for a schematic representation of the analysis design).
Methodological details on the analysis:
We applied principle coordinates analysis on a Jaccard distance matrix calculated for the entire community dataset. With redundancy analysis, we then tested the significance of the experimental treatments on the sample scores of the principle coordinates (Legendre and Anderson, 1999; Borcard et al.,
2011). We applied variation partitioning to estimate the amount of variation that can be explained by each of the experimental factors (Dispersal intensity,
Nutrient addition), their interaction (Nutrients x Dispersal) as well as the blocking factor (Inoculaton origin). The significance of the experimental factors was tested with 1000 random permutations. Db-RDA and variation partitioning were done in R (v2.13.0; R Foundation for Statistical Computing 2011), using the rda and varpart functions of the vegan package (Oksanen et al.,2005).
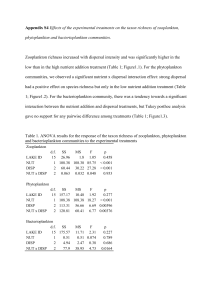
Table 4.1 RDA results for the effects of nutrient addition, dispersal intensity and inoculation origin on prokaryote and virus community composition. R 2 : fraction of explained variation, HNUT: high nutrient addition, LNUT: low nutrient addition, NNUT: no nutrient addition. Inoculation origin refers to the rows in the scheme of Appendix 1.
HNUT versus LNUT NNUT versus LNUT
Factor Covariable R 2 (%) F P R 2 (%) F P
Bacteria
Nutrient addition (Nutr) Disp, Inoc
Dispersal intensity (Disp) Nutr, Inoc
Nutr x Disp
Inoculation origin (Inoc)
Disp, Nutr, Inoc
Disp, Nutr
Viruses
Nutrient addition (Nutr) Disp, Inoc
Dispersal intensity (Disp) Nutr, Inoc
Nutr x Disp
Inoculation origin (Inoc)
Disp, Nutr, Inoc
Disp, Nutr
14.9
5.3
0
8
0
0
2
1.6
4.02
1.56
1.00
1.61
0.99
0.86
1.17
1.10
0.001
0.033
0.461
0.008
0.473
0.804
0.173
0.223
0
4
9
10
0
0.5
1.4
0
0.88
1.41
1.93
1.73
0.85
1.04
1.12
1.01
0.580
0.066
0.007
0.004
0.695
0.391
0.238
0.480
References
Borcard D, Gillet F, Legendre P (2011). Numerical Ecology with R. Springer: New York, NY.
Legendre P, Anderson MJ (1999). Distance-based redundancy analysis: testing multi-species responses in multi-factorial ecological experiments. Ecological
Monographs 69: 1-24.
Oksanen J (2005). Vegan: community ecology package. R package version 1.17-10.