SNP Genotyping Service Submission PLATFORM This document
advertisement

SNP Genotyping Service Submission PLATFORM
This document describes the procedure to follow when requesting a genotyping service using Sequenom
iPLEX Gold technology. Detailed instructions for the submission of samples and markers for genotyping
and information on the technology and its available products, on the genotyping procedures and on the
transmission of the results are also provided.
Sequenom iPLEX Gold Assay – Medium-Throughput Genotyping Technology
The Sequenom iPLEX Gold genotyping technology is ideal for medium sized projects (for SNP validation
or fine-mapping studies) when scoring between 5 and 400 bi-allelic markers such as SNPs (Single
Nucleotide Polymorphisms) and indels (insertion/deletion polymorphisms) on hundreds to a few
thousands of DNA samples is needed. A major advantage with this technology is that it is highly flexible
since there are no SNP type restrictions for the construction of the panel. It also allows the genotyping of
varied types of polymorphisms in any organism. The panel can contain up to 36 markers (for more details
see: Ehrich et al., 2005 Nucl Acids Res 33:e38).
The genotyping reaction is based on a multiplex PCR followed by a template-directed single base
extension using probes of various sizes. The products are then separated and detected by mass
spectrometry (MALDI TOF MS).
The assay conversion rate is between 85% depending on the projects. The call rate is about 92% and the
error rate is less than 0.1 %.
(주)바이오니아
49-3, Munpeong-Dong, Daedok-Gu, Daejeon 306-220, Korea
Phone: +82-42-936-8596 Fax: +82-42-930-8600
www.bioneer.com
info@bioneer.co.kr
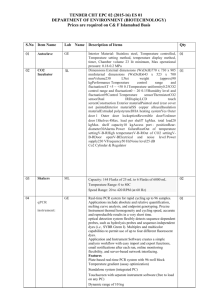
Submission Requirments
1. DNA Samples
To ensure the preservation of the DNA integrity during the shipment and the storage, please follow
these instructions. Do not send the samples in microtubes or in strip-tubes! The DNA samples must be
sent in 96-well plates using the recommended format types.
Plate Format
The DNA samples must be sent in 96-well plates in the appropriate plate type.
Plate Sealing
The DNA plates should be properly sealed. To seal full-skirted or half-skirted plates, use a good quality
adhesive film. For deep-well plates, use a compatible sealing mat.
Sample Shipment
The samples should be shipped frozen on dry ice and the DNA plates placed in a resealable zipper
plastic bag (Ziploc type).
Plate volume
Please, do not completely fill the wells with samples because this might increase the risk of samples
contamination. Use full-skirted or half-skirted PCR plates for low volumes (less than 50 μL) and use
deep-well plates for higher volumes (50 μL and more).
DNA Quality & Concentration
A minimum volume of 30 μL at a 20 ng/μL concentration should be sent for each sample. This is
enough for genotyping your samples with up to 4 Sequenom panels.
Minimum Processing Scale
Note that multiples of 96 samples are required for medium-throughput Sequenom iPLEX Gold
genotyping. These will be charged per multiple of 96 samples (whatever the total number of samples is).
* Recommended 96-Well Plate Types
Half-Skirt PCR Plates
- Any other Any half-skirt 96-well PCR plate
Full-Skirt PCR Plates
- Microseal PCR plates 96-well clear (Bio-Rad, cat# MSP9601)
- Any other full-skirt 96-well PCR plate
Adhesive Seals for Full-Skirt and Half-Skirt PCR Plates
- MicroAmp Clear Adhesive Films (Applied Biosystems, cat# 4306311)
- MicroSeal ’F’ Foil (Bio-Rad, cat# MSF-1001)
* Forbidden Plate Types
96-well cell culture plate
No-skirt 96-well PCR plate
(주)바이오니아
49-3, Munpeong-Dong, Daedok-Gu, Daejeon 306-220, Korea
Phone: +82-42-936-8596 Fax: +82-42-930-8600
www.bioneer.com
info@bioneer.co.kr
* DNA Plates Shipment
SNP Genotyping Service Submission sheet : Please fill out the SNP Genotyping Service Submission
sheet. Please send both an electronic and a hard copy of the SNP Genotyping Service Submission file
along with the shipment of DNA plates.
Samples : The samples should be shipped frozen on dry ice and the DNA plates should be properly
sealed and placed in a resealable zipper plastic bag (Ziploc type).
Client Management Office
Yong-Keun Jang, M.S.
Supervisor
Customer & Technical Support Team
49-3, Munpyeong-dong,
Daedeok-gu, Daejeon 306-220
Republic of Korea
Phone : 82-42-930-8650
Fax : 82-42-930-8600
Email : jyk2210@bioneer.co.kr
2. Marker
For the Sequenom iPLEX Gold technology, the panel design is done in-house.
No external design will be accepted. All panels must be ordered through us.
Typable Markers
Only bi-allelic markers such as SNPs (Single Nucleotide Polymorphisms) and indels (insertion/deletion
polymorphisms) with a single localization may be genotyped. There is no restriction on the SNP type.
Markers List Length
Since some markers will fail at the design level, it is recommended to provide a longer list in order to
have enough markers for the final constitution of the panel.
Flanking Sequence
Provide at least 200 base pairs of flanking sequence on each side of the marker.
Designability
Note that depending on the compatibility between the markers in a panel or on the marker context
sequence (the presence of repeats or low complexity sequences) it may be difficult to design
appropriate assays for some markers. Customers may be asked to replace some of them before
proceeding with the order of the custom panels.
(주)바이오니아
49-3, Munpeong-Dong, Daedok-Gu, Daejeon 306-220, Korea
Phone: +82-42-936-8596 Fax: +82-42-930-8600
www.bioneer.com
info@bioneer.co.kr
Target SNP
To specify the target SNP in the submitted sequence, put square brackets around the polymorphic locus,
and separate the alleles with a forward slash.
Target SNP Example: …TGC[A/C]CCG…
Target Indel
To specify the target indel in the submitted sequence, put square brackets around the polymorphic
locus, and separate the two alleles with a forward slash while specifying the indel with a minus sign.
Target Indel Example: …TGC[-/AT]CCG…
Lower-Case Masking
Repetitive or duplicated regions in the flanking sequence should be masked with lower-case nucleotides,
except for the 25 base pairs surrounding the targeted loci on each side.
Unmasked Sequence Example: …GTGACGAAATTTAAATTATTTTAAATTTAGCGT…
Masked Sequence Example:…GTGACGaaatttaaattattttaaatttaGCGT…
Neighbour SNPs and MNPs
Identify neighbour SNPs and MNPs in your sequence using the IUPAC (International Union of Pure and
Applied Chemistry) symbols listed in the Table 1.
Neighbour SNP Example: …GTTTGAA{G/C}CTGGACC… → …GTTTGAASCTGGACC…
Neighbour MNP Example: …GCGTGCAGT{C/G/T}CGCAGTGAC… → …GCGTGCAGTBCGCAGTGAC…
Neighbour Indels
Insertion/deletion polymorphisms (indels) surrounding the targeted loci should be indicated with an “N”.
Neighbour Indel Example: …GGACCA{-/CT}GGTTCGT → GGACCANGGTTCGT
* Table 1. International Union of Pure and Applied Chemistry (IUPAC) Symbols
IUPAC Symbols
Wobble Mixtures
R
A/G
Y
C/T
M
A/C
K
G/T
S
C/G
W
A/T
B
C/G/T
D
A/G/T
H
A/C/T
(주)바이오니아
49-3, Munpeong-Dong, Daedok-Gu, Daejeon 306-220, Korea
Phone: +82-42-936-8596 Fax: +82-42-930-8600
www.bioneer.com
info@bioneer.co.kr
V
A/C/G
N
A/C/G/T
* Formatted Sequence Example
Locus_Name: AB1234567
Formatted Sequence:
TTGAASCTGGACCANGGTTCGTCACACGTGGTTATACTGTTGGTTGTATTTATTATATAAATACATTTGATGATTCCCAA
CAGTTCTGGAGAACCTGC[G/T]TAATAATTGCTCAAAGCTAGAAATTGCATTCTCTAAAAACACAGCTAATCAATACA
ATATAGAGCCYaaaattttttaaatatatataataattttaaat
3. Validation Step
The validation is the first step in our Sequenom iPLEX Gold genotyping protocol. The aim is to assess
how assays do perform and if the quality of the DNA is high enough for the genotyping process. Once
the appropriate genotyping conditions for each panel are found the panel will be validated. Based on
the initial performance of each panel, the customer may decide to replace some markers (new panel
design and validation) or agree to continue with the same panel.
Panel and Assay Validation on Samples
One of your DNA plates will be used to test each panel (PCR and single-base extension). The first set of
genotypes obtained from this step is called Validation Genotypes. If good quality data are obtained,
with the customer's agreement, the rest of your plates will be processed as part of the production step.
Panel Redesign
The customer will be informed of the performance (success or failure) of each assay and panel. The
customer may decide to replace some markers or to design new assays for the failed markers. In that
case, the newly created panel will need to be validated as there is no guarantee, due to the dynamic
nature of multiplexing, that the markers that were functional in previous pools will still be successful in
the new ones. Each validation run should be approved by the customer and every validation experiment
will be charged.
4. Genotyping Lead Time
It usually takes from 3 to 4 weeks to receive all the assays and reagents. For projects of 6 plates and
less, usually 3 to 4 weeks are necessary to produce the genotypes and analyze the results. For larger
projects, it can take a little longer but customers will be informed of the progress.
(주)바이오니아
49-3, Munpeong-Dong, Daedok-Gu, Daejeon 306-220, Korea
Phone: +82-42-936-8596 Fax: +82-42-930-8600
www.bioneer.com
info@bioneer.co.kr
5. Results Transmission Modes
Transmission of Results by E-mail
6. Payment
A purchase order number (PO#) or an indication of payment by credit card should be provided before
starting any project. Do not provide your number, it will be asked later.
(주)바이오니아
49-3, Munpeong-Dong, Daedok-Gu, Daejeon 306-220, Korea
Phone: +82-42-936-8596 Fax: +82-42-930-8600
www.bioneer.com
info@bioneer.co.kr