Isolation of Total RNA from Yeast
advertisement

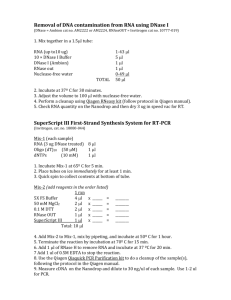
Cell and Molecular Biology Laboratory BIOL 406 1 Isolation of Total RNA from Yeast (This protocol revised&adapted from Dr. Karen Bernd, at Davidson College, N.C.) Objectives: 1. To learn how to work with yeast and extract Total RNA from them. 2. Understand the importance of RNases and how to protect RNA samples. 3. Quantitate and check purity of nucleic acid samples 4. Understand the difference between fermentation and respiration in yeast 5. Understand the steps required in experimental design and planning READ THIS PROTOCOL CAREFULLY. MAKE SURE YOU ARE PREPARED TO ASK ANY TECHNICAL OR THEORETICAL QUESTIONS BEFORE WE BEGIN THIS EXPERIMENT. IF IN DOUBT ABOUT ANYTHING, ASK! Isolation of total RNA from yeast RNA expression is one way of measuring gene activity. As genes are turned on, mRNA levels for those genes increase, generally resulting in increased protein expression. While cells have many ways of regulating proteins, differences in the functional state of the cell correlates directly with changes in mRNA levels. The cells use these ‘new’ proteins to respond to the current environmental challenges within the cell. In this experiment, we will monitor the yeast’s response to decreasing concentrations of glucose in the media as the yeast are forced to live under less favorable conditions. In rich media (i.e. high in glucose) the yeast metabolize the sugars to ethanol through anaerobic fermentation, but under less favorable conditions (little or no glucose) , they start metabolizing ethanol by aerobic respiration. This process of changing ‘food sources’ in yeast is called the diauxic shift. During the diauxic shift the yeast undergo substantial changes in gene expression related to the many biochemical pathways involved in the different methods of metabolism. We will try to gain insights into gene expression by examining total mRNA levels in yeast at critical time points throughout the shift using microarray technology. The first step in this process is to obtain yeast cultures at time points that represent the changes that occur during the diauxic shift, starting with yeast that have been grown in the presence of high glucose (i.e. the positive control) and subsequently in lower glucose concentrations. Your job is to isolate the total RNA at one time point. We will share our data, to get a Cell and Molecular Biology Laboratory BIOL 406 2 complete time course of the diauxic shift when we complete the microarray analysis for each time point later in the semester. In order to characterize the diauxic shift in yeast, you will be isolating total RNA from cultures of YEAST (STRAIN DBY10009) –a generous gift from the Botstein Lab, Princeton University, grown for different lengths of time . This particular strain of yeast has been completely sequenced. Later on, we will analyze gene expression of the ~6000 genes in yeast and try to correlate differences with their growing conditions. Your results for the future microarray work will depend on how well you complete this initial phase of the experiment. Make sure you work CAREFULLY. The most important step in RNA isolation is to remove as many sources of RNases from your work area as possible. RNases are enzymes whose entire purpose is to degrade RNA. They are very stable, low molecular weight proteins that can withstand high temperature and they are EVERYWHERE (yes, be paranoid!) ANY contamination with RNase will destroy your sample and pretty much wreck your day, experimentally speaking. RNA is extremely sensitive to degradation by RNases. How carefully you handle your samples and transfer solutions will have a huge impact on your yield. You must get rid of RNases. Your objective is to isolate a large quantity (perhaps 100 whole micrograms) of high molecular weight, undegraded total RNA. Read all instructions before touching anything. Make sure you have cleaned your bench space before beginning. Extra caution now will save you much work later. The main source of RNase contamination is your hands. Since it is out of the question to bake your hands at 210° for 15 hours to inactivate the enzyme, you must do everything you can to eliminate contact between your skin, things your skin has touched, and your precious samples. Clear your bench of all but the bare essentials. Once you have broken open the cells the RNAs are released and are susceptible to degradation by RNAases. Wear gloves and wash down your entire area with 'RNase ZAP' (special detergent that helps control RNases a bit.) All microfuge tubes and pipet tips have only been touched with gloves and have been sterilized extensively. The protocol is not difficult but you must be organized and careful. Cell and Molecular Biology Laboratory BIOL 406 3 Cell and Molecular Biology Laboratory BIOL 406 4 A. Materials: Yeast (Saccharomyces cerevisiae – Strain DBY10009) YPD Media (autoclaved) 10 g/L Yeast Extract 20 g/L Peptone 20 g/L Dextrose Lyticase (20 Units/mg) (store @ 4ºC, incubate @ 35ºC) (lyticase is an enzyme that breaks down yeast cells walls) BME (ß-mercaptoethanol) RNAse ZAP 100% ethanol Use ACS grade 100% ethanol or better. You will need 40 mL to add to the Wash Solution 2/3 Concentrate, and 1.25 mL per sample for use in step D.3. 15 ml tubes with tight fitting lids (ie corning) 50 ml Conical tubes 1.6 ml Eppendorf tubes 200 ul & 1000 ul RNase Free pipet tips Pipetmen 1.2 M Sorbitol, 10 mM KPhos pH 7.2 Potassium Phosphate (mw = 136.09 g/mol) Sorbitol buffer (mw = 182.17 g/mol) Ambion RiboPure – Yeast Kit (Cat #AM1926) Ambion RiboPure™-Yeast is a rapid RNA isolation kit that combines disruption of yeast cells with Zirconia Beads, phenol extraction of the lysate, and glass-fiber filter purification of the RNA. It can be used to isolate total RNA from a variety of yeast species. RiboPure-Yeast was extensively tested with Saccharomyces cerevisiae, Schizosaccharomyces pombe, Pichia pastoris,and Candida albicans. Amount Component 50 50 100 1.5 mL Screw Cap Tubes Filter Cartridges Collection Tubes (2 mL) 24 mL 2.4 mL 24 mL 95 mL 35 mL 50 mL Lysis Buffer 10% SDS* Phenol:Chloroform:IAA † Binding Buffer ‡ Wash Solution 1‡* Wash Solution 2/3 Concentrate Add 40 mL 100% ethanol before use Storage room temp room temp room temp 4°C 4°C 4°C 4°C 4°C 4°C Cell and Molecular Biology Laboratory BIOL 406 5 mL Elution Solution 80 g 550 µL 200 µL 550 µL Zirconia Beads (0.5 mm dia) 10X DNase 1 Buffer DNase 1 DNase Inactivation Reagent 5 4°C –20°C –20°C –20°C –20°C * If a precipitate develops during storage, dissolve the precipitate before use by warming the solution to 37°C. † Phenol:Chloroform:IAA is a poison and an irritant. Contact with it will cause burns. Use gloves and other personal protection equipment when working with this reagent. ‡ This reagent contains guanidinium thiocyanate, a potentially hazardous substance; use with appropriate caution. Vortex Mixer w/ adapter (Cat Ambion #AM10024) 95ºC Heating Blocks filled with deionized water 30ºC & 37ºC Water Bath Incubators Swinging bucket centrifuge @ 4ºC (G-309) 1.2% Agarose Gels DNA Gel boxes 10X Nucleic Acid Sample loading dye HyperLadder I Wide Range Markers Gel Running Bufffer (1 liter electrophoresis buffer 1x TBE) Ethidium Bromide Power Supply NOTES: Lab bench and pipettors Before working with RNA, it is always a good idea to clean the lab bench and pipettors with an RNase decontamination solution (e.g. Ambion RNase Zap Solution). Gloves and RNase-free technique • Wear laboratory powder free gloves at all times during this procedure and change them frequently. They will protect you from the reagents, and they will protect the RNA from nucleases that are present on skin. • Use RNase-free tips to handle the wash solutions and the Elution Solution, and NEVER put used tips into the kit reagents. Keep reagents on ice Always close pipet tip boxes Dispose of tips in waste containers, not on the bench Cell and Molecular Biology Laboratory BIOL 406 6 Never use tips more than once! B. Preparing yeast cells by making spheroplasts Yeast cells are grown in YPD media at 30º C in a shaking incubator. Yeast cultures grow exponentially until the media is exhausted. Growth of the yeast cultures is monitored by optical density, (Absorbance at 600 nm). Yeast are grown very similarly to bacteria. Like bacteria, they have different stages of growth. Scientists divide these stages of growth into early-log phase, mid log-phase, late log-phase, and the stationary phase. These phases are based on the amount of yeast that are in the broth and how quickly they are able to grow and divide based on nutrient use. Reading the absorbance of the sample at 600 nm is a way in which you can determine which phase of the yeast growth cycle you are in and how many cells you have. Here is a chart that will show you how this relates to yeast growth. You should record the O.D. of the class cultures. Early log-phase mid log-phase late log-phase stationary phase OD 600 cells/ml < 0.4 0.4 - 1.7 1.7 -6.6 > 6.6 < 107 1-5 x 107 5 x107 - 2 x 108 2 x 108 Haploid yeast cells contain approximately 1.2 pg of RNA per cell. The volume of culture for each time point was adjusted so that each sample (time point) contains the same amount of yeast. The cultures were inoculated and grown for 9 hours, then samples were withdrawn from the culture every 4 hours. The yeast cells were pelleted and then flash frozen and stored at -80ºC. Your group will receive yeast cells that were grown for a certain period of time (9 hrs, 13 hrs, 17 hrs or 21 hrs). We expect gene expression in the yeast to change because glucose was depleted over the time course. Yeast contain a hard cell wall. To improve our RNA yields we need to breakdown the cell wall so that the cells may be lysed more easily. Spheroplasts are yeast cells where the cell wall has been enzymatically degraded (ie using Lyticase). Cell and Molecular Biology Laboratory BIOL 406 7 1) Each group receives a yeast cell pellet that has been collected at one of the above time points (9 hrs, 13 hrs, 17 hrs or 21 hrs). 2) Label your yeast sample according to the time point you’re given so that you can identify your tube. Samples were standardized to contain 21 OD600 units. (i.e. 30 ml x 0.7 OD600 Absorbance units) Predict approximately how much RNA you should isolate. 3) Get your supplies ready: yellow and blue tips for pipetmen, 1.6 ml Microfuge tubes, and Potassium Phosphate/Sorbitol buffer. The next 3 steps are called a wash--they serve to move the frozen cells into a solution that is correctly buffered for the next procedure. 4) Add 1.0 ml of Potassium Phosphate/Sorbitol buffer to your yeast pellet. Let the yeast thaw on ice for at least 5 minutes. Resuspend the pellet by gently pipetting. 5) Label one 1.6 ml (large) eppendorf tube. Transfer the cells from the 15 ml conical tubes to the eppendorf tube. 6) Place the eppendorf tubes in the microcentrifuge (balancing tubes). Spin at 11,200g for 3min @ 4ºC. 7) Pour off supernatant (without disturbing pellet). Remove any residual supernatant with a pipetman. Resuspend pellet in 1.0 ml Potassium Phosphate/Sorbitol buffer. 8) Take tubes to hood. Add: 3µl ß-mercaptoethanol (a reducing agent that smells very bad) 320 µl of lyticase (enzyme that degrades cell wall components) 9) Place tubes in the incubator at 30°C (check TEMP!!) for 15min. The cells are now spheroplasts. Without a cell wall they are alive but structurally much weaker. Care must be taken so that the cells don't burst before you want them to. (How could the buffer they are in help keep them intact?) NOTE: yeast will repair their cell wall if the enzyme is removed so we cannot just keep a stock of 'wall free' yeast. Cell and Molecular Biology Laboratory BIOL 406 8 You must now get the cells ready for the next set of steps by washing away the lyticase and ß-mercaptoethanol. 10) Spin in microcentrifuge at 5600 g. Remove supernatant by pipeting (discard in hood to contain the BME smell). 11) Add 500µl potassium phosphate/sorbitol buffer. Repeat step 10 (one time). 12) While centrifuging make sure your ice bucket contains: 1 tube (screw cap) containing glass (zirconia) beads 1 tube of Lysis Buffer 1 tube of 10% SDS (Warm @ 37C to dissolve any precipitate) 1 tube of Phenol:Chloroform:IAA Clean RNA isolation area with 'RNAse ZAP' solution (1 spray bottle for lab) Isolating RNA The RiboPure-Yeast method disrupts yeast cell walls by beating cells mixed with an aqueous lysis buffer, SDS, phenol and 0.5 mm Zirconia Beads on a vortex adaptor (e.g. Ambion Cat #AM10024) for 10 min. Up to 3 x 108 cells can be processed at a time. The lysate is centrifuged to separate the aqueous phase, which contains the RNA, from the lower organic phase, which contains proteins, polysaccharides, and other cellular debris. The aqueous lysate is diluted with Binding Buffer and ethanol, and is drawn through a glass-fiber filter, which immobilizes the RNA. Contaminants are washed from the filter, and finally the RNA is eluted in a low ionic strength solution. Residual DNA is removed by treating the RNA with Ambion DNAfree ™ reagents (DNAse enzymes), included in the kit. As with all glass-fiber filter purification methods, 5S ribosomal RNAs and tRNAs are not quantitatively recovered using the RiboPure-Yeast Kit. The entire RNA isolation procedure and DNase treatment require approximately 1.5 hrs, starting with fresh, snap-frozen cultured yeast cells. The resulting RNA should be of superb quality, free of DNA and proteins, and suitable for use in virtually any downstream application. FROM THIS POINT ON DO NOT TOUCH ANYTHING ON YOUR BENCH WITHOUT WEARING GLOVES. DO NOT 'DOUBLETOUCH' TIPS DO NOT WALK AROUND WITH OPEN TUBES DO NOT LAUGH, TALK, OR BREATHE OVER OPEN TUBES (To quote the movies "be afraid--be very afraid") Cell and Molecular Biology Laboratory BIOL 406 9 C. Cell Disruption and Initial RNA Purification 1. Resuspend cells in lysis reagents Add the following to your yeast cell pellet in the order shown, and resuspend by vortexing vigorously for 10–15 seconds: (CAUTION: these reagent are extremely caustic; handle with respect) Amount Component 480 µL Lysis Buffer 48 µL 10% SDS (Check to make sure no precipitate is present) 480 µL Phenol:Chloroform:IAA 6 2. Add mixture to Zirconia Beads Transfer the mixture of cells and lysis reagents to the prepared tube containing ~750 µL cold Zirconia Beads, and securely fasten the lid. 3. Beat cells for 10 min on vortex mixer with a vortex adapter Position the sample tubes horizontally on the vortex adapter with the tube caps towards the center. Turn the vortex mixer on at maximum speed and beat for 10 min to lyse the yeast cells. 4. Centrifuge the lysed cells for 5 min at room temp, then transfer the aqueous phase to the 15 mL tube that contains 1.9 ml of binding buffer as explained below. a. Centrifuge for 5 min at 16,100xg at room temp to separate the aqueous phase, containing the RNA, from the organic phase. b. Transfer the aqueous phase (top), containing the partially purified RNA, to the 15 mL tube labeled “Binding Buffer”. Typically, the recovered volume from the cell lysis will be approximately 530 µL. D. Final RNA Purification 1. Before you start: Check Wash Solution 1 and redissolve by heating to 37°C if necessary A precipitate may form in the Wash Solution 1. Examine the reagents carefully before use, and if a precipitate is visible, redissolve it by heating the solution(s) to 37°C, and agitating the tube occasionally. Preheat Elution Solution to ~95°C Place 50 µL Elution Solution per sample into the heat block set to 95–100°C. The preheated Elution Solution will be used in step D.8. Inspect the Filter Cartridges Briefly inspect your Filter Cartridge before use. Occasionally, the glass fiber filters may become dislodged during shipping. If this is the case, gently push the filter down to the bottom of the cartridge using the wide end of a RNase-free pipette tip. NOTE: All centrifugation steps should be done at an RCF of ~13,000–16,000xg. Cell and Molecular Biology Laboratory BIOL 406 10 This is typically near the maximum speed setting on a microfuge. 2. Your sample now has a volume of ~2.4 ml (~530µL recovered from the lysis step and 1.9 ml of Binding Buffer.) This corresponds to 350 µL Binding Buffer for each 100 µL of partially purified RNA. 3. Add 1.25 mL 100% ethanol to your sample Add 1.25 mL 100% ethanol and mix thoroughly. (This corresponds to 235 µL ethanol for each 100 µL of partially purified RNA.) 4. Draw sample through a Filter Cartridge a. Apply 700 µL of the sample mixture to a Filter Cartridge assembled in a Collection Tube. The total volume of the lysate/ethanol mixture is ~3.7 mL, since the Filter Cartridge can only accommodate 700 µL, you will need to draw the lysate mixture through the Filter Cartridge in several applications of 700 µL at a time. b. Centrifuge for ~0.5–1 min or until the lysate/ethanol mixture is through the filter. c. Discard the flow-through and return the Filter Cartridge to the Collection Tube. d. Repeat as necessary to pass the entire sample through the Filter Cartridge. NOTE: The RNA is now bound to the filter in the Filter Cartridge. 5. Wash filter with 700 µL Wash Solution 1 a. Wash the filter by adding 700 µL Wash Solution 1 to the Filter Cartridge, and centrifuging for ~1 min or until all of the liquid is through the filter. b. Discard the flow-through and return the Filter Cartridge to the same Collection Tube. 6. Wash filter with 2x500 µL Wash Solution 2/3 a. Wash the filter by adding 500 µL Wash Solution 2/3 to the Filter Cartridge. Draw it through the filter as in the previous step. b. Repeat with a second 500 µL aliquot of Wash Solution 2/3. 7. Centrifuge for 1 min to remove excess wash solution from the filter a. Centrifuge the Filter Cartridge for 1 min to remove excess wash. b. Transfer the Filter Cartridge to a fresh 2 mL Collection Tube. 8. Elute RNA in 2x25 µL preheated Elution Solution a. Elute RNA by applying 25 µL Elution Solution, preheated to 95–100°C, to the center of the filter. b. Centrifuge for 1 min. c. Repeat the elution step with a second 25 µL aliquot of preheated Elution Solution into the same Collection Tube. Make sure your RNA is clearly labeled with your name, date, sample info etc. This a good stopping point, so we will freeze our samples until next week to finished the purification and analysis. STOP and Store samples at -70 ºC -- Cell and Molecular Biology Laboratory BIOL 406 11 Continue here at the next lab. E. DNase I Treatment RNA isolated using RiboPure-Yeast will contain contaminating chromosomal DNA. To remove it, we recommend including this DNase 1 treatment. The DNase Treatment and Removal Reagents provided with the RiboPure-Yeast Kit include ultrapure DNase I and reaction buffer that are rigorously tested to be sure that no RNase is present. This procedure makes use of a simple process to inactivate the DNase without the risk incurred by heating the RNA, or the hassles of adding EDTA or extracting with phenol/chloroform. 1. Assemble the DNase digestion reaction Assemble the DNase digestion reaction at room temp. Amount 50 µL 5 µ (1/10th vol) 4 µL Component RNA sample (from step D.8) 10X DNase 1 Buffer (i.e. 5–10 µL) DNase 1 (8 UNITS) 2. Incubate 30 min at 37°C Incubate the DNase digestion reaction for 30 min at 37°C. 3. Treat with 0.1 volume DNase Inactivation Reagent a. Resuspend the DNase Inactivation Reagent by flicking or vortexing the tube, then add 0.1 volume DNase Inactivation Reagent to each sample. For a typical 59 µL DNase digestion reaction, use 6 µL DNase Inactivation Reagent. b. Mix by vortexing and allow reactions to stand 5 min at room temp. c. Centrifuge 2–3 min at top speed in a microfuge (≥10,000 x g) at room temp to pellet the DNase Inactivation Reagent. Transfer the RNA (supernatant) to a fresh tube. THIS IS YOUR RNA. Place the tube on ice! Your RNA should always be stored on ice or frozen to slow degradation (why would cold slow degradation?) Question: What types of RNA have you just isolated? (how many different kinds of RNA are in a cell?) Cell and Molecular Biology Laboratory BIOL 406 12 F. RNA Storage RNA samples can be stored at –20°C for up to 1–2 months, or at –80°C for longer than 2 months. Checking RNA Integrity The quality of your RNA can be checked by agarose gel electrophoresis Prepare a 5 ug sample of your RNA for analysis by 1.2% Agarose gel. Coordinate with other groups to load samples (one sample from each time point) and markers on the gel. Run the gel at 120 mV for approximately 20 minutes until the two markers divide the gel into roughly equal thirds Stain the gel with ethidium bromide. Note the position of bands on the gel and the intensity of the staining. (Does your sample look like the ones shown the sample gel?) 1 = Degraded RNA 2 = Good RNA 3 = Good RNA