CRISPRCas9 FAQs
advertisement

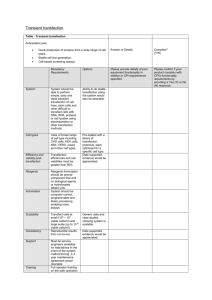
CRISPRCas9 FAQS 1.What are the benefits of using two RNAs (tracrRNA and crRNA) compared to a single guide RNA (sgRNA) expressed from a vector? The main benefit of using Edit-R with 2 synthetic RNAs is that RNAs for multiple targets can be ordered and transfected within a few days. This allows for rapid evaluation of multiple target sites (by testing multiple crRNAs) within a gene or editing multiple genes. CRISPR-Cas9 systems which utilize expressed guide RNAs require individual cloning of each targeting sequence into a vector backbone, which is especially time consuming when editing of several genes and several target sites per gene are required. 2. Will blasticidin selection help with my editing efficiency? We have found that if you have good transfection of Edit-R components that blasticidin selection only show^66666modest delivery of Edit-R components,antibiotic selection can improve efficiency. 3. What endotoxin testing method do you use for Edit-R? We use the LAL test from Charles River Laboratory. We certify that there is < 0.1 EU per µg of plasmid. 4. My DharmaFECT Duo transfection reagent arrived and the ice was melted. Is the reagent still ok to use? All DharmaFECT formulations, including DharmaFECT Duo reagent, have undergone substantial stability and functional testing, showing excellent performance even following storage at less-than-ideal conditions. The results are detailed in a product bulletin available for download. 5. Delivery of my package was delayed and my Edit-R Cas9 Nuclease Expression plasmid, mKate2 Transfection Optimization plasmid, tracrRNA and/or crRNAs have been at room temperature for a week. Are they OK to use? Samples are shipped as a dried pellet and are stable for two to four weeks at room temperature. All RNA oligonucleotides and plasmid DNA should be placed at -20℃ or -80℃upon receipt. 6.Dried RNA oligonucleotide pellets are stable at room temperature for two to four weeks, but should be placed at -20℃ or -80℃ for long-term storage. Under these conditions, the dried tracrRNA and crRNAs will be stable for at least one year. Maintaining We recommend not exceeding four to five freeze-thaw cycles to ensure product integrity 7.In what systems can I use the Edit-R kit? You can use the Edit-R in any mammalian system that is transfectable and where the hCMV promoter for Cas9 is active. 8. What is the formula for spectrophotometric quantification of RNA? To quantify RNA, use Beer's Law: Absorbance (260 nm) = (ε)(concentration)(path length in cm), where ε, epsilon, is the molar extinction coefficient (provided on the Product Transfer Form supplied with the order). When solved for the unknown, the equation becomes: Concentration = (Absorbance, 260 nm) / [(ε)(path length in cm)]. When a standard 10 mm cuvette is used, the path length variable in this equation is 1. If a different size of cuvette is used, e.g., a 2 mm microcuvette, then the path length variable is 0.2. 9. I'm using Edit-R and I see a fair amount of cell death after transfection of my cells. What can I do about this? Extensive cell death following transfection is an indication that delivery conditions need to be further optimized. Basic parameters to consider when optimizing transfections include transfection reagent and cell-specific conditions such as the amount of transfection reagent, the lot/batch of transfection reagent, duration of transfection, cell passage number and cell density at transfection. Often decreasing the amount of lipid present during transfection and/or the total duration of transfection will help minimize the toxic effect to the cells. Additionally, it is not uncommon to observe some variability from one tube of transfection reagent to another, and this may also represent a source of experimental variability. If the problem persists, we recommend that other transfection reagents be considered or you may contact Technical Support (ts.dharmacon@ge.com) for additional troubleshooting help 10.What quality control testing is performed for the synthetic tracrRNA and crRNAs? The Edit-R tracrRNA is purified by HPLC and evaluated by MALDI-TOF mass spectrometry and HPLC to confirm the length and purity. Custom crRNAs are evaluated with MALDI-TOF mass spectrometry. All synthetic RNAs are quantified by readings at A260 using a spectrophotometer. 11. I resuspended my crRNA in a buffer and there is a slight yellow tint to the solution. Is there something wrong? No. Deprotection of the bases during oligonucleotide synthesis uses a dithiolate derivative. Sometimes small quantities of this material remain in the sample (thus the yellow tint), but it will have no significant effect on editing experiments or cell viability. 12. I have read that shorter crRNAs (in the targeting sequence, from the 5′ end) are more specific. Can I order shorter crRNAs with less than 20 nucleotides of target sequence? We have validated the Edit-R system, as well as our manufacturing processes, for crRNAs with 20 nucleotides of target sequence. If a researcher wants to test a shorter crRNA (similar to those described in described in Fu, 2014), then a separate custom RNA oligo must be ordered through the custom single-stranded RNA synthesis route, the pricing will be very different, and we will not troubleshoot or support the results. 13. When should I use siRNA, shRNA, or Edit-R to study my gene of interest? For transient knockdown of a gene, siRNA is appropriate. For stable knockdown of a gene or where you would like to track stably-transduced cells using fluorescence, use shRNA. Both of these RNAi technologies cause knockdown of gene expression at the mRNA level. Edit-R is best for applications where a clonal line containing complete disruption and knockout of a gene at the DNA level is required. 14. Do crRNAs synthesized using Dharmacon 2'-ACE chemistry need to be HPLC-purified for gene editing in vitro? The proprietary Dharmacon 2'-ACE RNA synthesis chemistry has very high coupling efficiencies resulting in RNA oligos of exceptional yield and crude purity. Further crRNA purification is not necessary for gene editing experiments. Internal testing and successful gene editing were performed using desalted and deprotected crRNAs. 15. How should I store the Edit-R Cas9 Nuclease Expression and mKate2 Transfection Optimization plasmids? Dried plasmid DNA and DNA resuspended in buffer should be stored at -20 °C or -80 °C. Multiple freeze-thaw cycles should be avoided and strict DNase- and RNase-free conditions should be maintained to prevent degradation of nucleic acids. 16. What is the stability of the Edit-R tracrRNA and crRNAs? Dried RNA oligonucleotide pellets are stable at room temperature for two to four weeks, but should be placed at -20 °C or -80 °C for long-term storage. Under these conditions, the dried tracrRNA and crRNAs will be stable for at least one year. Maintaining sterile, RNase- and DNase-free conditions is always recommended as a critical precaution 17. Can I use the synthetic crRNA and tracrRNA components with other Cas9-expressing plasmids that are optimized for my biological system? The crRNAs and tracrRNA have been tested only with the Edit-R Cas9 expression plasmid in mammalian cell lines. However, the repeat component of the crRNA and the entire tracrRNA are sequences derived from S. pyogenes, so in theory these RNAs can be used with a S. pyogenes Cas9-expressing plasmid optimized for other biological systems (with an optimal promoter and codon usage for Cas9 expression). We cannot assure or guarantee any results. 18. How are the components of the Edit-R Gene Engineering System shipped? The Edit-R Cas9 Nuclease Expression plasmid DNA, mKate2 Transfection Optimization plasmid DNA, tracrRNA and crRNAs are all provided as dried pellets and shipped at room temperature. 19. My DNA mismatch assay shows a low percentage of editing. Why is that? DNA mismatch assays (e.g., SURVEYOR™, T7E1) rely on PCR amplification of a genomic DNA target site and observation of cleavage by a mismatch endonuclease, usually on a gel. While these assays are a straightforward approach for testing for editing, they likely underestimate editing for the following reasons: 1) Longer periods of mismatch enzyme digestion lead to off-target cuts that degrade the PCR product and reduce the intensity of the specific cut bands. 2) Cas9 cleavage and NHEJ can result in deletions, insertions, and mutations of various sizes; mismatch endonuclease cleavage may result in smeared bands that are not easily visualized on a gel. 3) If editing impacts primer binding (large inserts or deletions), the editing will not be detected. Observation of any cut bands in the DNA mismatch assay indicate successful editing, usually equal or greater than 1-10%. 20. Will blasticidin selection help with my editing efficiency? We have found that if you have good transfection of Edit-R components that blasticidin selection only shows a small improvement in editing efficiency. However, if you have modest delivery of Edit-R components, antibiotic selection can improve efficiency. 21. Do I have to use Cas9:tracrRNA:crRNA with DharmaFECT Duo Transfection Reagent? This system has been tested for use with DharmaFECT Duo, however, other reagents suitable for co-transfection of plasmid and RNA could be utilized, provided transfection optimization is performed. We cannot speak to any results obtained using other transfection reagents, nor can we provide any troubleshooting for poor transfection efficiency or cellular toxicity observed when other reagents are utilized. 22. Delivery of my package was delayed and the RNA oligos have been at room temperature for a week. Are they still fine? Samples are shipped as a dried pellet and are stable for 2-4 weeks at room temperature. All RNA oligos and siRNA should be placed at -20°C or -70°C upon receipt. 23. How are the components of the Edit-R Gene Engineering System shipped? The Edit-R Cas9 Nuclease Expression plasmid DNA, mKate2 Transfection Optimization plasmid DNA, tracrRNA and crRNAs are all provided as dried pellets and shipped at room temperature.