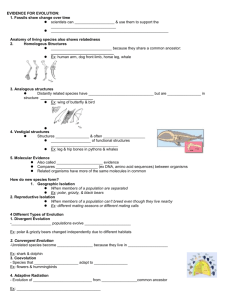
L318 – Evolution (Frey)
advertisement

POPULATION GENETICS SIMULATIONS By F. Frey and C. Lively, Indiana University How to get the program Go to http://evolution.gs.washington.edu/popgen/popg.html (This is correct for 2008. Use a browser other than Safari). Download the appropriate version of the popgen program for your computer. Software Notes See “Running the program” in the url above Purpose of this exercise Drawing from past courses and interactions with other faculty (especially Dr. Fortier and Dr. Dudle), Frank Frey and C. Lively developed this series of exercises. This program allows you to set parameter values (fitness of genotypes, mutation & migration rates, population size, etc.) and watch allele frequencies change through time in a series of simulated populations. These exercises should solidify your understanding of how selection, mutation, migration, and drift affect the evolutionary process. Additionally, you should come away with an understanding of how beneficial or deleterious recessive alleles may or may not persist in a population depending on the evolutionary forces at work and understand the effects of interacting evolutionary forces on the evolutionary process. How to set values in the program and start a simulation In the menu, select run, then select “new run”. With the mouse, click on the box neighboring the value you would like to set (or use the TAB button) Type in the new value. When all values are set, click the ‘run simulation’ button to start the simulation Parameters and explanations (alleles are italicized in this exercise) Initial frequency of allele A (not necessarily dominant – depends on relative fitness values) wAA, wAa, waa: Relative fitness of each genotype in the population (may vary between 0 & 1) Migration rate: Number of migrants from source population per generation to your population Mutation rate of allele A to allele a. This may vary between 0 & 1 Mutation rate of allele a to allele A. This may vary between 0 & 1 Number of different populations to simulate at the same time (may vary between 1 & 10) Number of generations the simulation will run through (may vary between 1 & 10000) Population Size: Size of each simulated population (may vary between 1 & 10000) 1 NATURAL SELECTION Consider a simple case of overdominance alone (no other evolutionary forces) using the parameters: p = 0.01, wAA = 0.9, wAa = 1.0, and waa = 0.9, where p is the frequency of the A allele. 1) Why is this a case of overdominance? 2) Will allele A get more or less common through time? 3) Will allele A be fixed (p = 1), lost (p = 0), or maintained at some intermediate frequency? 4) On the figure below, graph your prediction of the frequency of allele A through time. Consider the shape of the curve as you do so (e.g., Is the spread or loss of the allele constant? Does the rate of spread or loss of the allele slow down or speed up over time?, etc.). Is the output from Popgen, the dashed line is the analytical prediction, assuming an infinite population size. 1 Freq A 0 Time (generations) Simulation In the popgen simulation, set the number of populations to 1. Set Population Size to 10000 (so the effect of drift is small). Set the initial frequency of the A allele to 0.01. Set the mutation and migration values to zero. Now set wAA = 0.9, wAa = 1, waa = 0.9. Set the number of generations to 200. Click Okay to run simulation. 5) Do your predictions above match the results of the popgen simulation? 6) In the space below, summarize how overdominance alone affects the evolutionary process. 2 Now consider a case where allele A is recessive to allele a. Set p = 0.5 and waa = 1.0. All other parameters remain the same. 7) How do the three fitness values (wAA, wAa, waa) show that allele A is recessive? Is it a beneficial or deleterious allele? 8) Will allele A get more or less common through time? 9) Will allele A be fixed (p = 1), lost (p = 0), or maintained at some intermediate frequency? 10) On the figure below, graph your prediction of the frequency of allele A through time. Consider the shape of the curve as you do so (e.g., Is the spread or loss of the allele constant? Does the rate of spread or loss of the allele slow down or speed up over time?, etc.) 1 Freq A 0 Time (generations) Simulation Make sure p = 0.5, waa = 1.0, and all other parameters are the same as the previous simulation. Click on ‘run simulation’. 11) On the figure above, sketch the results of the simulation. Do your predictions match the results? 12) In the space below, summarize how recessiveness alone affects the spread or loss of an allele in the population. 13) How would you increase the strength of selection against allele A? Try simulating this situation. How does the strength of selection affect the rate of spread or loss of a recessive allele? 14) How could you change the genotype fitness values to make allele A a beneficial recessive allele? Try simulating this situation. Summarize the differences (at least 2) in evolutionary trajectory between a situation where allele A is a beneficial recessive and where allele A is a deleterious recessive. 3 MUTATION Consider a simple case of mutation alone. Things will happen extremely slowly if we use realistic mutation rates, so pretend our study population is at Love Canal, Chernobyl, or Three Mile Island. Set u (Ato a) = 0.1, u (a to A) = 0.01, p = 0, wAA = wAa = waa = 1. All other parameters remain the same. 15) Is mutation from allele A to allele a deleterious, beneficial or neutral? 16) Is mutation from allele a to allele A deleterious, beneficial or neutral? 17) Will allele A get more or less common through time? 18) Will allele A be fixed (p = 1), lost (p = 0), or maintained at some intermediate frequency? 19) On the figure below, graph your prediction of the frequency of allele A through time. Consider the shape of the curve as you do so (e.g., Is the spread or loss of the allele constant? Does the rate of spread or loss of the allele slow down or speed up over time?, etc.) 1 Freq A 0 Time (generations) Simulation Make sure all parameter values are set as described above. Click on ‘run simulation’. 20) On the figure above, sketch the results of the simulation. Do your predictions match the results? Press control C to continue the simulation for another 200 generations. 21) In the space below, summarize how mutation alone affects the spread or loss of an allele in the population. 22) Try some more simulations with lower mutation rates. For example, try u (A to a) = 0.01 & u (a to A) = 0.001. How do decreased mutation rates affect the time it takes for an allele to reach an intermediate frequency? Compare the results of the mutation simulations to the selection simulations. Note that mutation alone is a very weak evolutionary force relative to natural selection. 4 NATURAL SELECTION AND MUTATION Even though mutation alone is a weak evolutionary force, mutation is an extremely important evolutionary force. These next two examples will illustrate the effects of mutation when combined with selection on the evolutionary process. CASE I Consider a case of mutation – selection balance and the loss of deleterious recessive alleles. Set p = 0.5, wAA = 1, wAa = 1, waa = 0.5, u (A to a) = 0.001, u (a to A) = 0. All other parameters remain the same as before. 23) Will allele A be fixed (p = 1), lost (p = 0), or maintained at some intermediate frequency? 24) On the figure below, graph your prediction of the frequency of allele A through time. Consider the shape of the curve as you do so (e.g., Is the spread or loss of the allele constant? Does the rate of spread or loss of the allele slow down or speed up over time?, etc.) 1 Freq A 0 Time (generations) Simulation Make sure all parameter values are set as described above. Click on ‘run simulation’. 25) On the figure above, sketch the results of the simulation. Do your predictions match the results? There are two possible reasons why allele A was not fixed in this population (note: these are not mutually exclusive). First, mutation from A to a may be recreating allele a faster than selection takes it out. Second, allele a is ‘completely recessive’. This means that heterozygotes do not suffer a fitness cost because they have one copy of allele a. These next two simulations will test two hypotheses for the maintenance of allele a in this population. Hypothesis I: Mutation prevents the rapid fixation of allele A 26) Set the mutation rates to zero, leaving all other parameters the same. Re-run the simulation and compare your results to the previous simulation (with mutation rate A to a = 0.001). Does mutation alone explain the maintenance of allele a in the population? Hypothesis II: Allele A is not fixed in the population because allele a is completely recessive 27) Leaving the mutation rates at zero, change wAa to 0.9. This makes allele a ‘partially dominant’, meaning that one copy of allele a has a small deleterious effect on the heterozygote. Re-run the simulation and compare your results to the above two simulations. Does the ‘complete recessiveness’ of allele a alone explain the maintenance of allele a in the population? 5 28) In terms of evolutionary dynamics, what is the difference between a ‘completely recessive’ allele and a ‘partially dominant’ allele? To understand the importance of mutation, reset the mutation rates to u (A to a) = 0.001 and u (a to A) = 0. Leave all other parameters the same (allele a is still ‘partially dominant’). Re-run the simulation. 29) Compare these results to the prior situation of ‘partial dominance’ and no mutation. How does mutation affect the evolutionary dynamics? Re-run several simulations altering the mutation rate. 30) Can mutation alone always prevent the rapid fixation of allele A? Reset wAA = 1, wAa = 1, and waa = 0.5 leaving all other parameters the same (note: allele a is now completely recessive). Re-run several simulations altering the mutation rate (leave u (a to A) = 0). 31) How does the mutation rate affect the equilibrium frequency of allele A? Remember, there is a simple equation that determines the equilibrium frequency of a completely recessive allele given a constant mutation rate and constant selection differential. 32) Set the genotype fitness values to reflect any selection coefficient you choose (be sure that allele a remains completely recessive and deleterious). Set the mutation rate, u (A to a), to any value you choose. Run the simulation and determine if the equilibrium frequency of allele a in the population seems to match the expectation given by the equation. Remember, the y-axis of the graph is the frequency of allele A. The frequency of allele a is simply (1 – frequency of allele A). CASE II Consider another case of mutation – selection balance and the spread of beneficial recessive alleles. Set the following parameters: p = 1.0, wAA = 0.5, wAa = 0.5, waa = 1.0, u (A to a) = 0.0001, u (a to A) = 0, Ngen = 1000, population size = 10000. How do the fitness values show that allele a is a beneficial recessive? 33) Will allele A get more or less common through time? 34) Will allele A be fixed (p = 1), lost (p = 0), or maintained at some intermediate frequency? 6 35) On the figure below, graph your prediction of the frequency of allele A through time. Consider the shape of the curve as you do so (e.g., Is the spread or loss of the allele constant? Does the rate of spread or loss of the allele slow down or speed up over time?, etc.) 1 Freq A 0 Time (generations) Simulation Make sure all parameter values are set as described above. Click on ‘run simulation’. 36) On the figure above, sketch the results of the simulation. Do your predictions match the results? 37) Why does it take allele a so long to make it into the population even though it is an extremely beneficial allele? Set wAa = 0.6. This makes allele a ‘partially dominant’ because one copy of allele a has a small beneficial effect on the heterozygote. All other parameters remain the same. Re-run the simulation. 38) What is the effect of ‘partial dominance’ compared to ‘complete recessiveness’ on the evolutionary dynamics when a beneficial recessive allele is considered? (how is this simulation different from the previous one) Three important things to notice from these mutation – selection exercises. Mutation can counter-balance selection. Equilibrium allele frequencies may change depending on the mutation rate and the strength of selection (Case I). Mutation provides the raw genetic variation that selection can act on (Case II). The degree to which an allele is dominant or recessive in a population (from completely dominant to completely recessive and everything in between) affects evolutionary dynamics. The ‘effectiveness’ of selection in eliminating or fixing alleles in a population depends not only on the strength of selection but also on how alleles are expressed in the heterozygote (degree of dominance) 7 GENETIC DRIFT Popgen allow you to examine up to 10 different populations in the same simulation. We now want to look at the differences among populations over time for different population sizes. Genetic drift (sampling error) should increase as population sizes get smaller. Set p = 0.5, wAA = wAa = waa = 1, mutation rates = 0, migration = 0, Ngens = 100. Number of populations =8. 39) Predict what will happen if the population size = 1000 (will allele frequencies change from their initial values)? Run the simulation. 40) Explain why the dashed line remains at p = 0.5 throughout the simulation and the eight lighter lines ‘wiggle’ around the dark line. 41) Next, we’ll reduce the population size by ½ over the course of 5 more simulations (500, 250, 125, 64, 32). Predict how these simulations will differ from one another. Run the simulation. 42) Notice how in each of the above simulations the dashed line (infinite population size) remains steady on p = 0.5. When the population sizes get smaller the frequency of allele A goes to either 0 or 1 in the 8 lighter, single-population lines. Is the rate of fixation or loss of allele A related to population size? 43) Is genetic drift a creative force (creates genetic variation) or a destructive force (reduces genetic variation) within a population? Next, we’ll look at the effect of population size on the spread of a beneficial, ‘completely recessive’ allele in the population. Set wAA = 1.0, wAa = 0.9, waa = 0.9 and p = 0.3. Leave all other parameters as before. 44) Set the population size = 1000. Roughly how long does it take allele A to fix in the population? 45) Reduce the population size to 400, 80, 40 and 20. Compared to the previous simulation, what effect does reducing population size have on the spread of allele A in the population? 46) Now run the same simulation 10 times with a population size of 20. Why doesn’t allele A always get fixed? 47) Now make allele A ‘partially dominant’ (e.g., set wAa = 0.95) and re-run the same simulation 10 times with a population size of 20 and compare the results to the previous simulation. How does changing the degree of dominance affect the spread of allele A in the population? 8