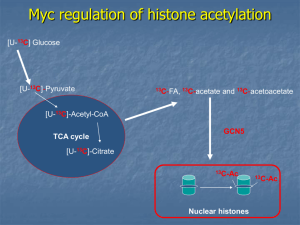
The c-myc gene-product, Myc, is a transcription factor that
advertisement

Supplementary Figure legends Figure S1 Gene expression and ChIP-seq data for the MDSR group of genes. (A) Differential transcriptional response of MDSR genes to serum stimulation in mycf/f;CreER cells, either without (orange bars) or with OHT to induce c-myc deletion (grey bars). The plotted values represent the fold-induction of each mRNA by serum, as determined from our microarray data. Only the maximal induction point during our time-course is reported for each gene. (B, C) Myc-binding within the TSS ±1kb interval for each MDSR gene in cells starved (red bars) or stimulated with serum for 8 hours (blue bars). The values represent Myc peaks in our ChIP-seq dataset, reported as OCR (B) or OCR-RMA (C). The dashed line in (C) indicates the OCR-RMA threshold of 15 (see text). Figure S2 Identification of regulatory patterns following Myc deletion and serum stimulation. The heatmaps show different classes of genes: (A) Up in rat, (B) Mycdependent repression, (C) Super-induced without Myc and (D) Known Myc-repressed genes (see text for definitions). The groups in (A, D) were determined based on previous literature and with the Myc database; those in (B, C) represent regulatory patterns in our microarray data, as those in Fig. 2. The genes in each group were clustered hierarchically on the basis of their microarray-based expression profiles. Changes in mRNA levels were expressed as fold-induction relative to time 0 in each culture, and color-coded as indicated. Figure S3 Quantitative RT-PCR validation of MISR and NRS genes. Cells were serum starved either untreated (black bars) or treated with OHT (grey bars) and then stimulated with serum for the indicated time. Average +/- sd of two independent experiments is shown for each gene. Figure S4 Quantitative RT-PCR validation of mRNAs expression. The figure shows the induction by serum (4 and 8h) in mycf/f;CreER cells, either without (blue bars) or with OHT pre-treatment to induce c-myc deletion (red bars). All data are normalized to the untreated control sample. The average +/- s.d. of three independent experiments is shown for each gene. (A) mRNAs included in the “Myc-dependent repression” group of Fig. S2B. The graphs are displayed in three sub-groups according to the outcome of the validation: Repressed and Myc-dependent, Repressed but not Mycdependent, Not repressed. (B) Gadd45a and Cdkn2b mRNAs (see text). Figure S5 Quantitative RT-PCR validation of mRNAs expression. The mRNAs shown are all from the group “super-induced without Myc” of Fig. S2B. The experiment was performed exactly as in Fig. S4. The graphs are displayed in two subgroups according to the outcome of the validation, as indicated. Figure S6 Correlation of Myc-binding measured by qChIP and ChIP-seq at 76 different loci in starved cells (top graph) or in cells stimulated with serum for 8 hours (bottom graph). Figure S7 Distribution of Myc peaks in the genome. (B) Absolute number of peaks in serum-stimulated cells, distributed over the window between -100 Kb and +100kb from TSSs. Each bin represents 1 Kb. (B) Distribution of intergenic Myc peaks relative to CAGE tags. Each intergenic Myc site has been annotated to the nearest Perna et al. 2010 p. 1 CAGE tag, along with a 10 times bigger dataset of randomly sampled intergenic regions showing the same length distribution. The two populations were then plotted as distributions relative to the nearest CAGE tag. The Myc intergenic sites display a sharper peak around zero. Figure S8 Myc-binding efficiency at promoters positively correlates with pre-existing mRNA levels. The box plots show the distribution of median mRNA levels in serumstarved cells, as determined from the microarray data, analysed for groups of genes bound by Myc with increasing efficiency following serum stimulation (defined by OCR-RMA values, as shown). Only Myc binding within the TSS ±1 kb window was considered. Perna et al. 2010 p. 2