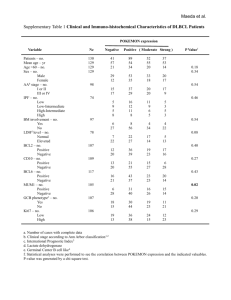
Supplementary Methods - Word file

Maeda et al.
Supplementary Methods
Retrovirus infection
For retrovirus-mediated gene transfer, Phoenix (ATCC) cells (3 x 10 6 ) were plated in a 10 cm poly-D-Lysine coated dish (BIOCOAT, BD) and, 16h later, cells were transfected with retroviral plasmid using Lipofectamine 2000 (Invitrogen). The virus-containing medium
(10 ml) was filtered and mixed with 5 ml of freshly prepared medium and supplemented with 4
g / ml polybrene (Sigma). 7 x 10 5 MEFs were plated in a 10 cm dish 16h before infection and viral supernatant was replaced on the day of infection. Antibiotics were administered 48h after infection and the cells were subsequently selected for 4 days and utilized for the various assays.
Real-time PCR analysis
Real-time PCR was carried out using QuantiTect SYBR Green RT-PCR Kit (Qiagen) and a Light Cycler (Roche). The primers used for p19 Arf were Arf-FW
(CGGAATCCTGGACCAGGTG) and Arf-Rev (ACCAGCGTGTCCAGGAAGC). For normalization, we utilized Rodent GAPDH control (ABI). To evaluate p14 ARF and
POKEMON mRNA levels in DLBCL cases, RNA was extracted from 37 cases of frozen
DLBCL samples using TRIZOL reagent (Invitrogen). Patient anonymity was insured and the study received a waiver by the Institutional Review Board (IRB). Porphobilinogen deaminasetubes ( PBGD ) gene was used for normalization and the following primers were utilized: p14 ARF -FW (AGCAGCCGCTTCCTAGAAGAC), p14 ARF -ReV
(CACGGGTCGGGTGAGAGT, as described 1 ), POKEMON -FW
(GAAGCCCTACGAGTGCAACATC), POKEMON -ReV
(GTGGTTCTTCAGGTCGTAGTTGTG), PBGD-FW (GGCAATGCGGCTGCAA) and
PBGD-ReV (GGGTACCCACGCGAATCAC).
ChIP assay
ChIP was performed utilizing a ChIP assay kit (Upstate) according to manufacturer’s specification with minor modifications. The polyclonal hamster anti-Pokemon antibody was raised against three different peptide sequences that are conserved among human and mouse ZBTB7 sequence. The primers for ChIP [Arf-ChIP-FW1
(GCTGGCTGTCACCGCGATG), Arf-ChIP-Rev1 (CGGCCTCACTGTGACAAGCG),
Arf-ChIP-FW2 (GAAGCGAGCGGGATCCGG) and Arf-ChIP-Rev2
(CGACCCATGCTCCCGCTG)] were designed to amplify a 296bp and a 302bp mouse p19 Arf promoter sequence respectively. The primers for
-actin
[
-actin-ChIP-FW
(TCGATATCCACGTGACATCCA) and
-actin-ChIP-Rev
(GCAGCATTTTTTTACCCCCTC)] were utilized as a control for the ChIP experiment.
PCR was performed using Platinum Taq DNA polymerase, Buffer A (Invitrogen) and 10% dimethyl sulfoxide (DMSO) with cycling parameters of 94ºC for 2 min, 32 cycles at 94ºC for 30 sec, 60ºC for 30 sec, 68ºC for 30 sec, and finally 68ºC for 3 min.
Pokemon mRNA expression level in transgenic founder lines
RNA was prepared from splenocytes using TRIZOL reagent (Invitrogen) followed by
Dnase I treatment (Invitrogen). cDNA was then generated utilizing a Su perScript™ III
System (Invitrogen) and PCR was subsequently performed with the following primer sets:
FLAG-Pok-FW ATTACAAGGATGACGATAAG, FLAG-Pok-Rev
CAGGATGTCGCCCACATTGG (for transgene specific FLAG-Pokemon), Pok-FW
GAGAAGAAAATCCGGGCCAAG, Pok-Rev GCAGCTATCGCACTGGTATGG (for total
Pokemon), hprt-FW CCTGCTGGATTACATTAAAGCACTG and hprt-Rev
GTCAAGGGCATATCCAACAACAAAC (for hprt).
Flow cytometry analysis
Flow cytometry analysis was performed according to a standard procedure using FITC
(B220, CD4 and CD44) and PE (CD3, CD8 and CD25) conjugated antibodies (BD) and a
FACScan analyzer (BD).
Immunohistochemistry for Human and Mouse paraffin embedded tissues.
Stains for POKEMON were performed with the anti-POKEMON hamster monoclonal antibody clone 13E9. If greater than 10% of malignant lymphoid cells showed moderate to strong nuclear positivity for POKEMON, then they were scored as positive. Nuclear intensity and percent positivity were also scored. Cases were further graded as: 0 = negative, 1 = weakly and moderately positive in greater than 10 of malignant cells and 2
= strongly and diffusely positive in greater than 50% of tumor cells. For Ki-67 analysis,
TMAs were scored as 0, no staining; 1+, less than 25% of tumor cells; 2+, 25-50%; 3+,
50-75%; 4+, >75% positivity in tumor cells. Ki-67 expression was analyzed as an ordinal variable, scored and grouped as 1, 2 vs. 3 and 4 for statistical purposes.
Pokemon stainings (with hamster monoclonal anti-Pokemon antibody, 13E9) were performed utilizing an automated staining machine (Discovery Staining Module, Ventana
Medical Systems Inc.) according to the manufacturer’s specification. For other molecules,
stainings were performed as follows. Slides were deparaffinized and hydrated to distilled water. Pretreatment protocols were used according to the primary antibody as follows:
BCL2 (monoclonal, DAKO Cytomation; citric acid, pH 6.0, 1:200 dilution); MUM1
(monoclonal, DAKO Cytomation; EDTA, pH 8.0, 1:100); CD10 and BCL6 (monoclonal,
Novocastra Laboratories; citric acid, pH 6.0, 1:100) were used for DLBCL samples. CD3
(polyclonal, DAKO Cytomation, Carpenteria; citric acid, pH 6.0, 1:10); Cytokeratin
Cocktail AE1:AE3 (monoclonal cocktail, BioGenex; with protease 24 (30min at 37 o C),
1:800); TdT (polyclonal, DAKO Cytomation; citric acid, pH 6.0, 1:40) were used to characterize mouse tumors. Endogenous peroxide was quenched with a 5 minute incubation in 3% hydrogen peroxide diluted in distilled water, washed in distilled water for
1 minute, then rinsed in PBS, pH 7.2 twice. Antibody dilutions were made in 1% bovine serum albumin (BSA)/phosphate buffered saline (PBS). Slides were incubated with the primary antibody overnight at 4 o C in a humidity chamber. Secondary antibody at 1:500 dilution in 1% BSA/PBS was applied for 1 hour at room temperature in a humidity chamber, then slides were washed three times in PBS for ten minutes each.
Peroxidase-conjugated streptavidin at 1:500 in 1% BSA/PBS was applied for 45-60 minutes at room temperature in a humidity chamber then washed three times in PBS for ten minutes each. Slides were transferred to a diaminobenzidine (DAB) bath dilute in
PBS for 5-15 minutes, washed in tap water for one minute, then counterstained using
Harris modified hematoxylin (Fisher), decolorized with 1% acid alcohol and blue in ammonia water, dehydrated three times each in 95% ethanol, 100% ethanol and xylene for two minutes each, then coverslipped with Permount mounting media.
Statistical analysis
A log-rank statistical analysis was utilized for overall survival (OS) outcome in DLBCL cases. Survival curves were calculated according to the Kaplan-Meier product limit method. Correlations between pathologic, clinical features and POKEMON expression were made using the chi-squared test for univariate variables. The R version 1.9.1 software (http://www.r-project.org/) was used for statistical analysis.
Reference
1. Linggi, B. et al. The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat
Med 8 , 743-50 (2002).