Labelling,Hybridization and Wash Protocol
advertisement

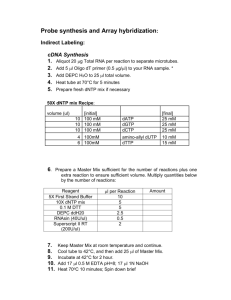
Microarray hybridization Protocol Adapted from TIGR and Keck Center Grace, June 03. Aminoallyl Labeling 1. Precipitate and dry in the air up to 200μg of total RNA which has been DNAse I treated, dissolve in 14.5μL RNase- free water. Add 2μg (4μL, 500ng/ μL) dT18 primer. 2. Mix well and incubate at 70°C for 10 minutes. 3. Place on wet ice and spin down. 4. Add: 5× First Strand Buffer 0.1M DTT 50× aminoallyl- dNTP mix SuperScript II RT (200U/ μL) RNAse inhibitor 6 μL 3 μL 0.6 μL 2 μL 20U, or 0.5 μL 5. Mix well and incubate at 42°C for 2 hours to overnight. 6. To hydrolyze RNA, add 2U (1μL, 2U/ μL) RNAse H, incubate at 37°C for 15 minutes. 7. Add 25 μL of 1M Tris pH 6.8 to neutralize pH. (This is important! DNA binds to Qiagen Column at pH below 7.5.) Reaction Purification I: Removal of unincorporated aa- dUTP and free amines (Qiagen PCR Purification Kit). 1. Mix cDNA reaction with 300 μL, or 5× Volume, buffer PB (Qiagen supplied) and transfer to QIAquick column. 2. Place the column in a 2ml collection tube (Qiagen supplied) and centrifuge at ~ 10,000g for 1 minute. Empty collection tube. 3. To wash, add ~750 μL phosphate wash buffer to the column and centrifuge at ~ 10,000g for 1 minute. Empty collection tube. 4. Repeat the wash step. 5. Empty collection tube and centrifuge column an additional 1 minute at maximum speed. 6. Transfer column to a new 1.5mL Eppendorf tube. 7. Carefully add 30 μL phosphate elution buffer to the center of the column membrane. 8. Incubate for 1 minute at room temperature. Elute by centrifugation at ~ 10,000g for 1 minute. 9. Elute for a second time into the same Eppendorf tube by repeating step 7 and 8. The final elution volume should be ~ 60 μL. 10. Dry the sample in a speed vac for about 1 hour. Coupling aa-cDNA to Cy Dye Ester. 1. Resuspend aminoallyl- labeled cDNA in 4.5 μL 0.1M sodium carbonate buffer pH9. 2. Add 4.5 μL of the NHS- ester Cy dye. 3. All reaction tubes should be wrapped in foil and keep in dark. Incubate for 1 hour at room temperature. Reaction Purification II: Removal of uncoupled dye (Qiagen PCR Purification Kit). 1. To the reaction add 35 μL 100mM NaOAc pH5.2. 2. Add 250 μL, or 5×Volume, buffer PB (Qiagen supplied) and transfer to QIAquick column. 3. Place the column in a 2ml collection tube (Qiagen supplied) and centrifuge at ~ 10,000g for 1 minute. Empty collection tube. 4. To wash, add ~750 μL buffer PE (supplied) to the column and centrifuge at ~ 10,000g for 1 minute. Empty collection tube. 5. Centrifuge column an additional 1 minute at maximum speed. 6. Transfer column to a new 1.5mL Eppendorf tube. 7. Carefully add 30 μL buffer EB (supplied) to the center of the column membrane. 8. Incubate for 1 minute at room temperature. Elute by centrifugation at ~ 10,000g for 1 minute. 9. Elute for a second time into the same Eppendorf tube by repeating step 7 and 8. The final elution volume should be ~ 60 μL. Analysis of Labeling Reaction. 1. Use a 50 μL Beckman quartz MicroCuvette to analyze the entire undiluted sample in a sepctrophotometer. 2. Wash the cuvette and dry. 3. Pipette sample into cuvette and measure absorbance at 260nm and either 550nm for Cy3, or 650nm for Cy5, as appropriate. 4. Pipette sample back into the original sample tube. 5. For each sample calculate the total picomoles of cDNA synthesized using: Pmol nucleotides = [OD260 * Volume (μL) * 37ng/ μL] / 0.3245ng/pmol 6. For each sample calculate the total picomoles of dye incorporation using: Pmol Cy3 = OD550* Volume (μL) / 0.15 Pmol Cy5 = OD650* Volume (μL) / 0.25 7. nucleotide / dye ratio = pmol cDNA / pmol Cy dye 8. > 200pmol of dye incorporation per sample and a ratio of between 20 to 90 nucleotides / dye molecules are optimal for hybridizations. 9. Mix the two differentially labeled probes which will be hybridized to a same microarray slide for study of relative gene expression. The final volume should be ~ 120 μL. 10. Dry the probe mixture in a speed vac at room temperature in dark. Rehydration of Microarray Slides, necessary for our Arabidopsis and Rice Arrays 1. Rehydrate the slides on ~ 40°C water (dots facing water) for 15 seconds. Use large beaker to ensure even moisturizing. 2. UV cross link (wavelength 1600) twice. 3. Wash the slides in 1% SDS by dipping in and out for 2 minutes. 4. Wash the slides in hot water (~ 95°C) by dipping in and out for 2 minutes. 5. Wash the slides in 95% Ethanol for 10 – 20 times. 6. Centrifugation at ~ 1000rpm for 2 minutes to dry the slides. 7. Store the slides in a dry and dark place; prepare for pre- hybridization. Prehybridization 1. Preheat 50mL tubes filled with prehybridization buffer in 42°C water bath. 2. Place slides in tubes, ensure that arrays are covered. 3. Incubate in 42°C water bath for 45 minutes, shaking occasionally. 4. Wash slides in two beakers of Millipore water thoroughly. 5. Wash slides in isopropanol. 6. Centrifugation at ~ 1000rpm for 2 minutes to dry the slides. 7. Hybridize immediately. Hybridization 1. Add ssDNA (final concentration 10 μg / ml) to probe and fill to 40 μL with sterile water. 2. Heat probe at 95°C for 2-3 minutes. 3. Place slide in hybridization chamber. 4. Add 40 μL of 2× hybridization buffer, mix well and pipette to slide. Cover carefully with coverslide. Close chamber. 5. Hybridize at 42°C for 1- 2 days. Keep the incubator moist. Wash 1. Wash the slides in tubs. First wash: 1× SSC, 0.2% SDS at 42°C for 4 minutes. Second wash: 0.1× SSC, 0.2% SDS at room temperature for 4 minutes. Third wash: 0.1× SSC at room temperature for 4 minutes. 2. Centrifugation at ~ 1000rpm for 2 minutes to dry the slides. Scan and Data analysis Solutions used: 1. 50× aminoallyl- dNTP mix: Aminoallyl dUTP: For a final concentration of 100mM add 19.1 μL of 0.1M KPO4 buffer pH7.5 to a stock vial containing 1mg of aa-dUTP. Vortex gently to mix; then transfer the aa-dUTP solution into a microfuge tube. Store at -20°C. Measure the concentration of the aa-dUTP solution by diluting an aliquot 1:5000 in 0.1M KPO4 buffer pH7.5 and measuring the OD289. Stock concentration in mM = OD289 * 704. Labeling Mix (50×) with 2:3 dUTP:dTTP ratio: Mix the following reagents: dATP(100mM) 5 μL dCTP(100mM) 5 μL dGTP(100mM) 5 μL dTTP(100mM) 3 μL aa-dUTP(100mM) 2 μL Total: 20μL Store the mix at -20°C. 2. 1M Tris pH 6.8, 100mM NaOAc pH5.2: Prepared according to Molecular Cloning. 3. Phosphate wash buffer, phosphate elution buffer: Prepare 1M K2HPO4 and 1M KH2PO4. To make 1M Phosphate buffer (KPO4) pH 8.7, mix: 1M K2HPO4 9.5mL 1M KH2PO4 0.5mL For 100mL Phosphate wash buffer (5mM KPO4, pH8.0, 80% Ethanol), mix: 1M KPO4, pH8.7 0.5mL Millipore water 15.25mL 95% Ethanol 84.25mL Note: wash buffer will be slightly cloudy. Phosphate elution buffer is made by diluting 1M KPO4 pH 8.7 to 4mM with Millipore water. 4. 0.1M sodium carbonate buffer pH9: Prepare 1M Na2CO3 buffer pH9.0: Dissolve 1.08g Na2CO3 in 8mL Millipore water; add 1.35mL HCl (12N); then add 0.75mL water. To make 0.1M Na2CO3 buffer, dilute 1:10 with water right before use. Note: Na2CO3 buffer changes composition over time; prepare it fresh every week. 5. NHS- ester Cy dye: Dilute one tube of dried product in 45 μL DMSO in dark, mix well and aliquot to 10 tubes. Wrap with foil and keep at -80°C. Use one aliquot for each sample. Always try to prevent photobleaching! 6. Prehybridization buffer: 20% Formamide 50mL Formamide 5× Denhardts 25mL 50× Denhardts 6× SSC 83mL 20× SSC 0.1% SDS 2.5mL 10% SDS 25 μg/mL tRNA 0.625mL 10mg / mL tRNA Millipore water 86mL Total: 250mL 7. 2× hybridization buffer: 50% Formamide 5mL Formamide 10× SSC 5mL 20× SSC 0.2% SDS 0.2mL 10% SDS Total: 10mL 8. 20× SSC: Dissolve 17.53g NaCl, and 8.82g Sodium Citrate in 80mL Millipore water. Adjust pH to 7.0 with 12N HCl. Bring final volume to 100mL with water. Sterilize by autoclaving.