BL5203 - Department of Biological Sciences
advertisement
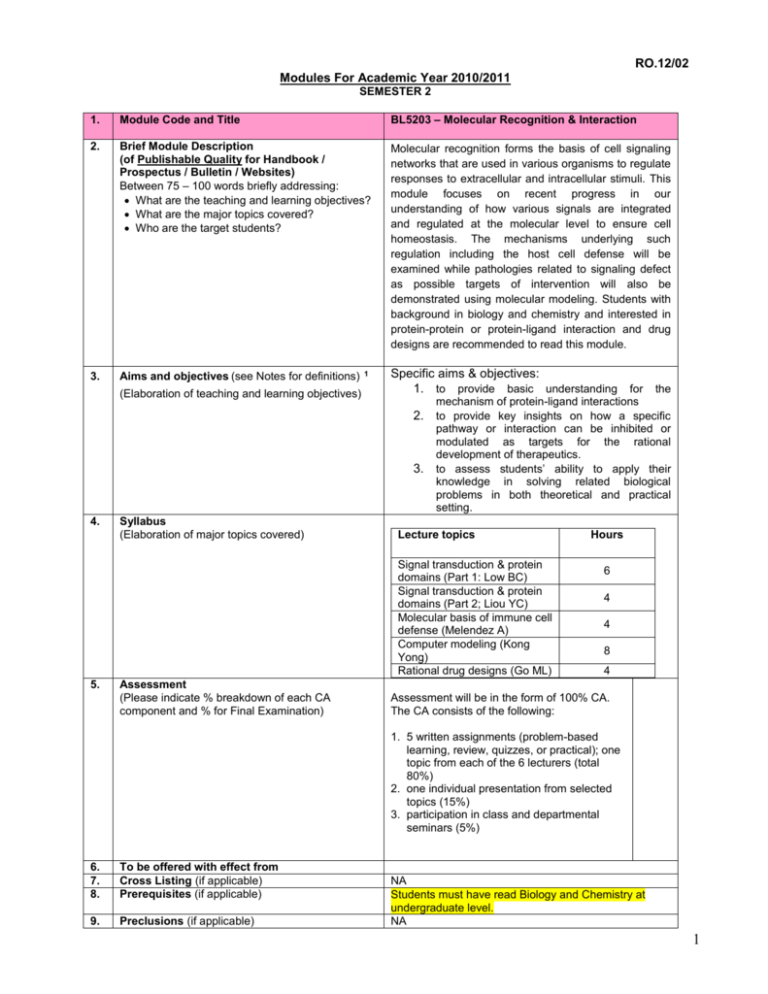
RO.12/02 Modules For Academic Year 2010/2011 SEMESTER 2 1. Module Code and Title BL5203 – Molecular Recognition & Interaction 2. Brief Module Description (of Publishable Quality for Handbook / Prospectus / Bulletin / Websites) Between 75 – 100 words briefly addressing: What are the teaching and learning objectives? What are the major topics covered? Who are the target students? Molecular recognition forms the basis of cell signaling networks that are used in various organisms to regulate responses to extracellular and intracellular stimuli. This module focuses on recent progress in our understanding of how various signals are integrated and regulated at the molecular level to ensure cell homeostasis. The mechanisms underlying such regulation including the host cell defense will be examined while pathologies related to signaling defect as possible targets of intervention will also be demonstrated using molecular modeling. Students with background in biology and chemistry and interested in protein-protein or protein-ligand interaction and drug designs are recommended to read this module. 3. Aims and objectives (see Notes for definitions) (Elaboration of teaching and learning objectives) 1 Specific aims & objectives: 1. to provide basic understanding for the mechanism of protein-ligand interactions 2. to provide key insights on how a specific 3. 4. Syllabus (Elaboration of major topics covered) pathway or interaction can be inhibited or modulated as targets for the rational development of therapeutics. to assess students’ ability to apply their knowledge in solving related biological problems in both theoretical and practical setting. Lecture topics Signal transduction & protein domains (Part 1: Low BC) Signal transduction & protein domains (Part 2; Liou YC) Molecular basis of immune cell defense (Melendez A) Computer modeling (Kong Yong) Rational drug designs (Go ML) 5. Assessment (Please indicate % breakdown of each CA component and % for Final Examination) Hours 6 4 4 8 4 Assessment will be in the form of 100% CA. The CA consists of the following: 1. 5 written assignments (problem-based learning, review, quizzes, or practical); one topic from each of the 6 lecturers (total 80%) 2. one individual presentation from selected topics (15%) 3. participation in class and departmental seminars (5%) 6. 7. 8. To be offered with effect from Cross Listing (if applicable) Prerequisites (if applicable) 9. Preclusions (if applicable) NA Students must have read Biology and Chemistry at undergraduate level. NA 1 10. Module Lecturer(s) 2 Name(s)/Department(s): 11. 12. Modes of Teaching and Learning (Lectures, regular tests, Q & A, IVLE, problembased learning) Basic Reading List Compulsory reading Supplementary reading 13. 14. 15. Maximum Class Size Modular Credits (MC) Workload Per Week (The workload for a 4-MC module must add up to 10 hours per week. E.g. 2 hours lecture; 1 hour tutorial; 7 hours preparatory work) Name(s): Low BC (DBS)*, Liou YC (DBS), Chris Hogue (DBS), Liu DX (IMCB); Zhang ZL (IMCB) Email: dbslowbc Tel.No: 6516-7834 (Low BC; coordinator) This module comprises lecture series in various related areas of specialities. The assessments are based on either writing reviews, problem-based learning or practical on computational modeling. Some tutorials will be available. Students will also prepare presentation on selected topics to provide their critical evaluation and are strongly encouraged to participate in group discussions. Up-to-date scientific articles (reviews or research papers) are heavily used for both teaching and discussion. Extra reading materials will be provided when necessary but students are expected to undertake further literature reviews themselves 30 4 Lecture hours per week: 3 Tutorial hours per week: 1 Laboratory hours per week: 0 No. of hours per week for projects, fieldwork, 0 assignments, etc. : No. of hours per week for preparatory work 3: 6 Total hours per week: 10 Notes: (Please refer to Paper R.205/02A ‘Policy on Modules’) 1 2 3 Aims specify the knowledge and abilities we expect students to acquire. Objectives are expressed in operational terms and specify what the students will be able to do at the end of the module. For regular module, at least 2 regular faculty members should be qualified to teach. No. of hours of preparatory work is the time a student is expected to spend in preparing for tutorials, projects, assignments, etc. 2 BL 5203 Course contents A. Signal transduction Part 1 Low BC (6 h) Several types of signalling (proteins, ions/metabolites, lipid derivatives) and levels of signaling cascades (ligands/receptors, intracellular, nucleus) will be introduced with focus on small GTPases signaling and cell dynamics control. Regulation by protein-protein interaction will be discussed in the context of specificity, redundancy and cross-talks. Various protein-protein interaction domains, protein-lipid binding domains, recognition/structural and functional motifs, dockers, adaptors, scaffolds will also be introduced. Strategies using classical and contemporary biochemistry, pharmacological intervention, molecular biology and proteomic tools will be highlighted in appropriate contexts. B. Signal transduction Part 2 Liou YC (4h) C. Molecular basis of immune cell defense Bernard Leong (4 h) The topics will deal with how the immune system is orchestrated and how inappropriate regulation of certain signals could contribute to autoimmunity or the inability to mount effective immune responses against particular pathogens or cancers. Topics to be covered include the molecular recognition of antigenic peptides by the MHC molecules and howw the immune response can be triggered by the binding of peptides (self peptides an foreign peptides), intracellular signalling cascades triggered by various immune receptors (TCR, BCR, and FcRs), and chemokines, in different cell types. Will also focus on the pathways leading to the release of lipid-derived pro-inflammatory mediators, degranulation, cytokine production, and chemotaxis (including vascular-endothelial cell activation). D. Molecular Modeling Kong Yong (8 h including practicals) Two major components will be covered: 1. Computer modeling of molecular recognition: Molecular surface and mechanism of ligand binding. Computer modeling of molecular surface. From surface profile to cavity recognition. Representation of a cavity. Modeling of molecular recognition: Ligand-protein docking. Applications of ligand-protein docking in drug/ligand design Practicals: Ligand-protein docking software 2. Computer modeling of molecular interaction: Forces involved in molecular interaction. Potential energy representation of forces. Conformation optimization for molecular interaction. Modeling of conformation change due to molecular interaction. Applications of conformation optimization/change in drug/ligand design Practicals: Protein and ligand-protein structure optimization. F. Rational drug designs (Go ML 4 h total) Four major components will be covered: 1. Drug Targets: chemistry of drug binding to receptors; drug receptors and physiological mediators; receptors as primary targets of drug action: ligand gated ion channels, receptors with instrinsic enzymatic activity, G-protein coupled receptors; intracellular receptors; drugs acting through voltage sensitive ion channels (sodium, 3 calcium, potassium, chloride channels); drugs acting through transporters; Drug action mediated through enzymes; drugs influencing synaptic transmission 2. Drug design strategies: the lead compound and how are leads identified; criteria for lead compounds and how to optimize a lead compound: bioisosterism, conformational restraint, homologation, upsizing/downsizing approaches; physicochemical properties and drug design. 3. Peptide based drug-design: ligand-receptor interactions; conformational analysis of peptides in drug-design; docking; peptidomimetic design. 4. Examples of how drugs are designed/discovered: captopril : ACE inhibitor; Cimetidine: H2 antagonist; Losartan: angiotensin II antagonist 4