Supplemental Material and Methods (doc 70K)
advertisement

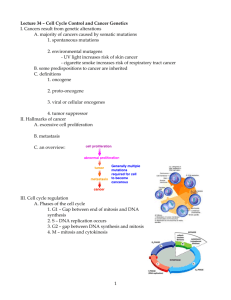
Detailed Materials and Methods: Cell Culture, Plasmids, and Transfections: MCF-7 and U2OS cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) at 37 °C in 5% CO2. DMEM was supplemented with 10% fetal bovine serum; 100 unit/ml penicillin-streptomycin, and 2mM L-glutamine. The GFP-cyclin D1a and D1b expression plasmids have been described previously (Solomon et al., 2003). Plasmids were transfected using the lipid-based transfection reagent Lipofectin (Invitrogen), in accordance with the manufacturer’s recommended protocol. Antibodies, Immunoblotting and Immunostaining: Antibodies against the following proteins were used: cyclin D1a (Ab-3; NeoMarkers), Lamin B (sc-6217; Santa Cruz), Cyclin D1b (Burd et al., 2006; Wang et al., 2008). Immunoblotting employed standard conditions For immunostaining cells were cultured on glass coverslips and fixed with 3.7% formaldehyde in PBS for 10 minutes. with 0.4%Triton X100 in PBS. Fixed cells were permeabilized for 15 minutes Subsequently cells were stained using the primary antibodies at 1:100 dilutions of the indicated antibody in IF (PBS, 3% BSA, 0.5% NP40) buffer, for 1 hour at 37 C. Coverslips were washed with PBS and subjected to Texas Red conjugated secondary antibody (1:500) in IF buffer for 1 hour. Coverslips were washed with PBS, counter-stained with DAPI, and mounted with gelvatol. Imaging was performed on a Zeiss Axiovert microscope equipped with Axiocam image capture software, the Texas Red and GFP images were captured at the same exposure condition. DAPI images were captured using automatic exposure settings. Case Control Study Population: The Multiethnic Cohort Study (MEC) consists of over 215,000 men and women in Hawaii and Los Angeles (with additional African-Americans from elsewhere in California) and has been described in detail elsewhere (Kolonel et al., 2000). The cohort is comprised predominantly of African Americans, Native Hawaiians, Japanese Americans, Latinos and European Americans who entered the study between 1993 and 1996 by completing a 26-page self-administered questionnaire that asked detailed information about dietary habits, demographic factors, personal behaviors, history of prior medical conditions, family history of common cancers, and for women, reproductive history and postmenopausal hormone use. The participants were between the ages 45 and 75 at enrollment. Incident cancers in the MEC are identified by cohort linkage to population-based cancer Surveillance, Epidemiology and End Results (SEER) registries covering Hawaii and Los Angeles County, and to the California State cancer registry covering all of California. Beginning in 1994, blood samples were collected from incident breast cancer cases and a random sample of MEC participants to serve as a control pool for genetic analyses in the cohort. Controls were frequency matched to cases based on race/ethnicity and age (in 5-year intervals). Eligible cases in this nested breast cancer case-control study consisted of women with incident breast cancer diagnosed after enrollment in the MEC through May 2002. Controls were women without breast cancer prior to entry into the cohort and without a diagnosis up to May 2002. The breast cancer case-control study in the MEC consists of 1,376 invasive breast cancer cases (287 African Americans, 360 Japanese, 281 Latinas, 92 Native Hawaiians, and 356 Whites) and 2,583 controls (673 African Americans, 429 Japanese, 707 Latinas, 311 Native Hawaiians, and 463 Whites). This study was approved by the Institutional Review Boards at the University of Southern California and at the University of Hawaii. SNP Genotyping: The G/A870 polymorphism was genotyped using TaqMan (Applied Biosystems, Foster City, CA) at the University of Southern California. Primers, probes, and assay conditions are available upon request. The genotyping success rate was 96%. Replicate blinded quality control samples (5%) were included to assess reproducibility of the genotyping procedure; the concordance was 100%. Automated quantitative analysis (AQUA) of immunohistochemical staining: Immunohistochemistry and AQUA analyses were performed on sections of a tissue array constructed by cutting edge matrix assembly (CEMA) (LeBaron et al., 2005) that contained 143 invasive breast carcinoma specimens and 37 normal breast tissues. Briefly, after deparaffinization and rehydration of array sections, antigen retrieval was performed by microwave treatment in citrate buffer, pH 6 (DAKO). Sections were blocked with 10% goat serum and followed by incubation of antibodies to either Cyclin D1a (Lab Vision, clone SP4; Cat# RM-9104) at a dilution of 1:100 or Cyclin D1b at a dilution of 1:50 for 1 h. Sections were then washed 3 times with TBS and subsequently incubated with a mouse cytokeratin antibody (DAKO, Cat# AE1/AE3) for 1 h. The Cyclin D1a and Cyclin D1b antibodies were detected using an anti-rabbit HRP-conjugated secondary antibody (DAKO EnVision-Plus), followed by incubation with Tyramide-Cy5 (Perkin Elmer, Cat# NEL745). Cytokeratin was visualized by further incubating the sections with a mouse secondary antibody conjugated to Alexa 488 (Molecular Probes, Cat# A11034). Finally, all sections were stained with DAPI (Vector, Cat: H1500) for nuclear visualization. Automated quantitative analysis was performed using the AQUA/PM2000 Imaging Platform (HistoRx) as described (Dolled-Filhart et al., 2006). Tissue array slides were scanned and images of each breast cancer tissue were captured at different channels detecting FITC/Alexa 488, Cy5, or DAPI. AQUA software was then used to identify epithelial masks based on FITC-positive cytokeratin- expressing cells. AQUA scores were calculated for Cyclin D1a and Cyclin D1b representing average signal intensities within epithelial cells. Statistical tests comparing mean levels of D1a/D1b in normal versus cancer were performed by Student’s T-test, with error bars representing 95% confidence intervals. p<0.05 was considered significant. Assessment of D1a/D1b levels versus grade of invasive cancer was performed by ANOVA with an additional LSD posthoc test. Error bars represent the 95% confidence interval, and p<0.05 was considered significant. Quantitative PCR (qPCR). RNA was isolated from tissue samples via standard Trizolbased methodologies. Subsequently, cDNA was generated using SperScript VILO (Invitrogen). qPCR was performed using a StepOne machine (Applied Biosystems). Express SYBR (Invitrogen) assays were used to determine GAPDH (F- CCAGGTGGTCTCCTCTGACTTC, R-TCATACCAGGAAATGAGCTTGACA), Cyclin D1a (F- CTCTCCAGAGTGATCAAGTGTGACCC, R- TGTGCAAGCCAGGTCCACC), and Cyclin D1b (F- AACACGCGCAGACCTTCGTT, R- TAAGCCCCGGCAAGGCT). Relative expression of cyclin D1a and cyclin D1b transcripts in tumor specimens relative to non-neoplastic control was determined as previously described (Pfaffl et al., 2002). Breast Cancer Outcome Series: Immunohistochemistry for cyclin D1a, cyclin D1b and Ki67 was performed on formalin-fixed paraffin-embedded tissue microarrays (TMA) containing 1mm diameter cores derived from 175 patients with invasive ductal carcinoma, treated at St Vincent’s Hospital, Sydney, Australia. Between 2-6 cores of tumor were present for each patient. Clinical follow-up was available for a median of 73.5 months (range 0-149 months). The clinicopathological details of this cohort are summarized in Supplementary Table II. Four m thick sections were mounted on Superfrost Plus adhesion slides (Lomb Scientific, Australia). Slides were baked at 78ºC before being deparaffinised in xylene and rehydrated through serial alcohol solutions (100%, 95%, 70%). Endogenous peroxidase activity was blocked using 3% H2O2 for 5 minutes, followed by protein block-serum free solution (DAKO Corporation Carpentaria, CA) for 30 minutes. Following antigen retrieval sections were incubated withthe following antibodies: cyclin D1a (clone SP4, Neomarkers/LabVision, Fremont, CA), cyclin D1b or Ki67 (clone SP6, Neomarkers/LabVision), followed by detection using Envision+ Rabbit (DAKO) and visualisation with DAB+ (DAKO). Sections were then counterstained with Mayer’s haematoxylin (DAKO), dehydrated and mounted for analysis. The negative control employed concentration matched non-specific rabbit IgG in place of the primary antibody. Stained slides were assessed by an experienced breast pathologist (EKAM) who was blinded to the clinicopathological parameters and patient outcome. Cores were scored for both the percentage and intensity of cells which showed nuclear staining: 0 negative, 1+ weak, 2+ moderate, 3+ strong. Positive staining was considered if present in >1% of cells. An average intensity was calculated from the scores obtained across all cores of tissue for each patient. A modified “Histo score” was also calculated by multiplying the intensity by the percentage of positively staining cell nuclei. Differences in patient numbers for assessment of different antigens was due to loss of cores during processing and staining of the TMAs. Statistical Analyses of Clinical Outcomes: Data was analysed using Statview 5.0 software (Abacus systems, Berkley CA) with a p value of <0.05 accepted as significant. Disease specific survival, defined as the date of definitive procedure until the date of death due to breast cancer, locoregional recurrence (ipsilateral breast, chest wall, axilla or supraclavicular fossa) and distant metastases (to liver, lungs, bones, brain or distant lymph nodes) were used to assess outcomes using Kaplan-Meier and Cox Proportional Hazards (for univariate and multivariate analyses respectively). There were a total of 51 metastastic cases, and 56 cases of recurrence (5 local and 51 metastasis). For cyclin D1a there is the convention that ~50% of tumors overexpress the protein and thus the data were dichotomized by the median H-score. Irrespective of this cutpoint the outcome data with cyclin D1a remain largely invariant. Given that this is the first study to relate cyclin D1b expression to outcome in human breast cancer there is no historically defined basis for dichotomizing the data. Therefore, serial cut-points with sequential Kaplan-Meier analyses (logrank test) to define between high and low expressing groups. This approach defined the cut point of intensity >2+ that divides the population in an approximate ratio 2:1 which is similar to the split between ER/PR positive and negative which are well established clinical prognostic/predictive biomarkers. Those factors that were prognostic on univariate analysis were then assessed in a multivariable model to identify factors that were independently prognostic. REFERENCES: Burd CJ, Petre CE, Morey LM, Wang Y, Revelo MP, Haiman CA et al (2006). Cyclin D1b variant influences prostate cancer growth through aberrant androgen receptor regulation. Proc Natl Acad Sci U S A 103: 2190-5. Dolled-Filhart M, McCabe A, Giltnane J, Cregger M, Camp RL, Rimm DL (2006). Quantitative in situ analysis of beta-catenin expression in breast cancer shows decreased expression is associated with poor outcome. Cancer Res 66: 5487-94. Kolonel LN, Henderson BE, Hankin JH, Nomura AM, Wilkens LR, Pike MC et al (2000). A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol 151: 346-57. LeBaron MJ, Crismon HR, Utama FE, Neilson LM, Sultan AS, Johnson KJ et al (2005). Ultrahigh density microarrays of solid samples. Nat Methods 2: 511-3. Pfaffl MW, Horgan GW, Dempfle L (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. Solomon DA, Wang Y, Fox SR, Lambeck TC, Giesting S, Lan Z et al (2003). Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. J Biol Chem 278: 30339-47. Wang Y, Dean JL, Millar EK, Tran TH, McNeil CM, Burd CJ et al (2008). Cyclin D1b is aberrantly regulated in response to therapeutic challenge and promotes resistance to estrogen antagonists. Cancer Res 68: 5628-38.