comparing quantitative trait loci and gene expression data
advertisement

COMPARING QUANTITATIVE TRAIT LOCI AND GENE
EXPRESSION DATA ASSOCIATED WITH A COMPLEX TRAIT
Bing Han*1, Naomi S. Altman*1, David J. Vandenbergh2, Jessica A. Mong34, Laura Cousino Klein25, Michele McClellan Stine2, Ryan
Peterson5, Donald W. Pfaff3
1
Department of Statistics, The Pennsylvania State University, University Park, PA, US
2
Department of Biobehavioral Health, The Pennsylvania State University, University Park, PA, US
3
The Laboratory of Neurobiology and Behavior, Rockefeller University,
4
Department of Pharmacology & Experimental Therapeutics, University of Maryland School of Medicine
5
Center for Developmental and Health Genetics, The Pennsylvania State University, University Park, PA, US
* To
whom correspondence should be addressed.
Abstract
We develop methods to compare the positions of quantitative trait loci (QTLs) and of selected sets of
genes. We apply our methods to QTLs for addictive behavior in mouse, and sets of genes associated in
microarray studies with the nucleus accumbens (NA) region of the brain. The link between the QTLs and
NA genes is moderately stronger than expected by chance. Statistical methodology developed for this study
can be applied to similar studies to assess the joint information in microarray and QTL analyses.
1 Introduction
The association between complex phenotypic trait and genetic markers on the chromosome can be
detected through statistical analysis, leading to the identification of QTLs – regions of the chromosome that
appear to be associated with the phenotype. QTLs are expected to be associated with the genes controlling
some aspect of the phenotype. One mechanism by which a gene might be associated with the trait is
through altered transcription. This transcriptional regulation is easily measured by microarray analysis.
Microarrays have the ability to measure all of the genes in the genome, which parallels the genome-wide
scan performed by QTL methods.
Several investigators have considered combining QTL and microarray data for studying a genetic trait.
For example, Wayne and Mclntyre (2002) proposed a way of identifying candidate genes based on both
QTL mapping and microarray data. Fischer et al. (2003) developed a web-based software tool for combined
visualization and exploration of gene expression data and QTLs. The methodology developed in this work
is complimentary to the analyses that can be performed on the GeneNetwork website (WebQTL,
www.genenetwork.org), which allows assessment of the relationship between gene expression and QTLs in
Recombinant Inbred mice (Wang et al., 2003).
However, comparing QTL and microarray data is not completely straightforward. First, the estimated
range of QTL positions is generally wide, containing thousands of putative genes. However, QTL analysis
may also miss some interesting genes (Wayne and Mclntyre, 2002). Second, the high level of experimental
error and limitations of analysis in microarray data introduce mistakes in the identification of relevant
genes.
Further problems arise when we try to associate phenotypes with gene expression in specific tissues.
While the association is direct if the tissue defines the phenotype, unanticipated associations can arise if the
tissue indirectly regulates the phenotype – for example, bone strength may be regulated through physical
activities regulated by the brain. Alternatively, association can arise through plieotropic expression of the
gene in a tissue not included in the expression study but in which the gene plays a role in the phenotype. In
addition, the association between a phenotype and a tissue may depend on ephemeral conditions that
may not be present when the tissue was collected for the microarray study or on a small percentage of cells
in the organism, which may be masked by bulk tissue preparation.
In this paper, we suggest several methods to examine the strength of association between a group of
QTLs and a set of genes identified from a microarray study. As a byproduct, the methods can also provide
information about the association between two traits or a trait and a tissue. We apply our methods to the set
of mouse QTLs identified from the literature and the sets of mouse genes identified from a microarray
study. First, we identified a set of 120 QTLs associated with drug abuse behaviors in mice (Jung, 2003)
from the Mouse Genome Informatics database (http://www.informatics.jax.org).
Gene expression data were derived from microarray analysis of RNA purified from brain regions of
one-day old mice. Male and female C57BL/6J pups from 4 litters were sacrificed approximately 6 hours
after birth. The brains were removed and placed on an ice-cold platform and bathed with ice-cold 0.1M
Phosphate Buffered Saline (PBS). Three coronal slabs containing the Basal Forebrain (BF, including the
Nucleus Accumbens), Preoptic Area (POA), and Medial Basal Hypothalamus (MBH), were isolated from
the brain by a series of cuts based on the anatomical description of GD18 mouse brain. All cuts were made
under a Zeiss dissecting microscope. The first coronal slab containing the BF corresponded to plates 8-9
and was made by placing the first cut 2-3mm caudal to the leading edge of the cortex and a second cut
1-1.5mm from the first.
The coronal section containing the POA corresponded to plates 10-12, and was
made by a third cut immediately in front of the optic chiasm or approximately 2.0 mm from the second cut.
Finally, the third slab contained the MBH, corresponding to plates 15-16, was cut from the brain by making
two cuts at the beginning and end of the median eminence, respectively. From the first tissue slab, a
rectangular block of tissue containing the BF was dissected by making vertical cuts immediately lateral to
the anterior commissure and two horizontal cuts, the first immediately dorsal of the anterior commissure
and the second approximately 0.5mm from the ventral surface. The POA was dissected from the second
slab in another rectangular block. Again, two vertical cuts were made immediately lateral to the ventricles
and one vertical cut was made immediately below the anterior commissure. Finally, a 2 mm trapezoid
containing the MBH was dissected from the coronal slab by making two diagonal cuts from the dorsal tip
of the third ventricle to the base of the brain and a third cut at the dorsal tip of the third ventricle and
parallel to the base of the brain. The tissue of interest was immediately placed in ice-chilled RNAlater
(Ambion, Inc., Austin, TX) and stored at –80ºC. RNA was isolated from the brain tissue by
homogenization in TRIZol (Invitrogen, Carlesbad, CA) following the manufacturers protocol. The RNA
pellet was dissolved in RNase free water and further purified using the RNeasy RNA purification kit
(Qiagen, Valencia, CA).
Approximately 41±6.4 (mean ± S.D.)
m the MBH.
Separate pools of RNA were created from 4
pups for each of the 3 brain regions, and 2 sexes. At least 3 separate litters were represented in each pool to
minimize possible litter-specific effects. Target cRNA was prepared for hybridization to microarray chips
following the manufacturer’s instructions (Expression Analysis Technical Manual, Affymetrix Inc, Santa
Clara, CA). Bacterial RNA purchased from Affymetrix was spiked into the RNA to serve as internal
controls. A portion of the cRNA was hybridized to Test Array 2 chips to determine quality of the cRNA,
and was followed by hybridization of the cRNA to 6 Murine Genome Array (MG-U74Av2) chips.
The
hybridization, washing, developing, and scanning of the chips were carried out following the Affymetrix
protocols. Raw signals from the chip were processed using Microarray Suite 5.0 (MAS, Affymetrix) and
internal controls were found to produce expected signals. All genes that received an “Absent” call by MAS
were discarded from subsequent analysis.
An average of each gene’s expression signal from the male and
female chips for each brain region was used to generate a ratio of the NAc to the POA, and of the NAc to
the MBH.
Those genes with a ratio of greater than 1.5 for both comparisons were selected as
NAc-enriched.
Of the 179 genes on this list, the Affymetrix ID numbers of five genes could not be
positioned on the mouse genome and may not be true genes. The resulting list of 166 genes that are
preferentially expressed in the NAc was used in the analysis of gene-QTL relationships described below.
The NA plays an important role in mouse behaviors relevant to drug abuse. We expect the strong
association between the QTLs and the NA genes.
2 Exploratry data analysis and quantification of link
Figures 1 shows the correspondence between the the set of QTLs and the set of NA genes. The long
horizontal dashed lines are numbered to represent the mouse chromosomes. Note Y chromosome is
apparently shorter than others and no data were available regarding gene expression or QTLs on it. The
short discrete horizontal segments are the spans of the QTLs defined as +/- 5 centiMorgans (cM) from the
peak position. The small circles in the center of every segment are the peak positions of the QTLs. Finally
the vertical lines are the NA genes. The data we work with are from Affymetrix®, but the plot is drawn
using the Bioconductor suite in R (Gentleman et al., 2004).
QTL and NA genes
Y
X
19
18
17
16
Chromosome
15
14
13
12
11
10
9
8
7
6
5
4
3
2
1
0.0 e+00
5.0 e+07
1.0 e+08
Basepairs
1.5 e+08
2.0 e+08
Fig. 1. Combined visualization of QTLs and NA genes
QTLs are measured in centiMorgans (cM), which measures recombination frequency between
markers on a chromosome. Gene locations are usually measured by the physical distance in base pairs (bp)
or megabase pairs (1 Mb =106 bp). Empirically, on average 2 Mb = 1 cM in the mouse chromosome. There
are a few more accurate methods to translate cM into Mb (e.g. Silver, 1995, Fischer et al, 2003, and Voigt
et al, 2004). To match QTL sets and gene sets, we need to measure locations on the same scale. We
adopted the embedded conversion tool in Expressionview (Fischer et al, 2003) to estimate physical
distances from cM. The “smoothing window” technique used in Expressionview essentially applies the idea
of piecewise regression. However at the edge of chromosomes and some middle places where possibly near
to the cutting points of the “smoothing windows”, we found that Expressionview gives apparent poor
estimations. In those cases we use polynomial regression to estimate physical distance from cM by using
genes for which both measures are available. This method also has good performance except at some ends
of a chromosome. Any QTL with a span that extends beyond the end of a chromosome is truncated. No
obvious matches between the QTL set and the NA genes can be seen Figures 1. The visual impression does
not support a strong association between them.
We consider two approaches to quantify the strength of a link. For convenience, we denote a set of
QTLs, such as drug abuse QTLs, by Q and a set of genes, such as the NA genes, by G. A natural first
approach is to consider the percentage of genes in G covered by the whole span of Q. The link between Q
and G is strong if this number is big. This quantification reflects the “completeness” of Q in terms of
covering G. This suggestion is supported by data from Drosophila, in which co-regulated genes are found
in clusters (Spellman et al., 2002). A second approach is to consider whether each QTL in Q covers at least
one gene in G. If a QTL in Q covers no genes in G, it is called “empty”; otherwise it is “non-empty”. The
link between Q and G is strong when the percentage of empty QTLs is small. This quantification reflects
the “accuracy” of Q in terms of covering G. If Q is strongly associated with G, we expect both
completeness and accuracy to be high. However the two methods do not necessarily give the same result
because they are measuring different sides of an association. In other word, an accurate QTL set could be
not complete, and vice versa. While each method can answer the question if the link is strong in terms of
completeness or accuracy, we want to develop a unique measure on both completeness and accuracy
together to answer the question: is the link strong? This is defined by a weighted average of completeness
and accuracy. Firstly we need to introduce a few notations. Let N be the number of genes in G, M be the
number of QTLs in Q, n be the number genes in G covered by Q, and m be the non-empty QTLs in Q
covering genes in G. So it is straightforward to define completeness C = n / N, and accuracy A = m / M. We
define the combined measure of a link as
S
C
A
M N
(1)
The weight is chosen to diminish the effect of “coincidence” or matching by chance. When M increases
more area of the genome will be covered by Q. Then the completeness C will also increase no matter
whether the underlying link is strong or not. To punish the effect of a big M, we use 1/M as the weight of
completeness. It is similar for the choice of weight on accuracy.
The limiting behaviors of the combined measure S satisfies the need to differentiate a strong link from
a “noised” link where matching primarily results from matching by coincidence. Let s be the number of
genes in G really having matching relationships with some QTL in Q. Correspondingly let r be the number
of QTLs in Q really matches some genes in G. Note r is not necessarily equal to s. Besides the true
matching relationship, every gene has a probability p = p(M) to be covered by Q. On the other hand every
QTL has a probability q = q(N) to be non-empty with respect to G. By introducing the new notation, the
completeness can be written as
s
I{gene is matched }
genes w/o
true match
C
N
.
(2)
Then the expectation of C is easy to write down
s ( N s) p
.
N
EC
(3)
Similarly
I{QTL is non - empty }
r
QTLs w/o
true match
A
EA
Then
,
(4)
M
ES
r ( M r )q
.
M
r s ( N s ) p ( M r )q
MN
(5)
(6)
Consider the following limiting circumstances: 1. (perfect match) when s → N and r → M, ES will
monotonically increases to the limit (M+N) / MN; 2. (totally random) when s → 0 and r → 0, ES will
monotonically decreases to the limit (Np+Mq) / MN; 3. (G mess up) when N → ∞ and fix M, notice q=q(N)
→ 1 in this case, ES will converge to p / M; 4. (Q mess up) when M → ∞ and fix N, notice p=p(M) → 1 in
this case, ES will converge to q / N. From the above it can be concluded that the combined measure S will
approach its maximum when a perfect match arises and decrease when the link weakens in some face.
3 Statistical tests for accuracy and completeness
Until the biology is fully understood, we cannot be certain if the link is truly random. In this section,
we determine the statistical significance of the observed levels of completeness and accuracy compared to
random association, by comparing with a null distribution determined by simulation. Random selection of
QTLs is not readily done as selection of random intervals along the chromosomes is unlikely to model the
true distribution of QTLs. However, since the physical locations of all genes on the microarray are known,
random sets of genes are readily created by choosing genes at random, and considering the completeness or
accuracy of the QTL sets with respect to these genes.
To assess the strength of association between a QTL set Q and a gene set G of size N, we compute the
completeness and accuracy of Q. We then select genes at random from all the genes represented on the
microarray. The simplest way to do this is to select N genes at random from the array (the unconditional
method). However, since there is considerable variability in the percentage of tissue-specific genes on each
chromosome, and since the QTLs may not be randomly distributed among chromosomes, we can also
consider selecting Ni genes from the ith chromosome, where Ni is the number of genes in the gene set on the
chromosome (the conditional method). By repeatedly selecting gene sets at random and computing the
completeness and accuracy for Q, a null distribution (unconditional or conditional) is computed. The
p-value for the observed completeness or accuracy is the percentage of simulated data sets for which the
completeness (accuracy) is as strong as or stronger than the observed value. The estimated p-values are
displayed in Table 1 based on 1,000 random rounds.
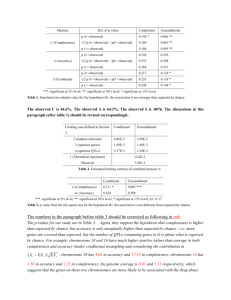
Table 1. Simulated one-sided p-value for the hypothesis H0: the link is not stronger than expected by chance.
Measure
Def. of p-value
C (Completeness)
A (Accuracy)
S (Combined)
Conditional
Unconditional
p (# >observed)
0.085 **
0.045 ***
p (1/2 # observed + # >observed)
0.103 *
0.053 **
p (>= observed)
0.120
0.060 **
p (# >observed)
0.192
0.151
p (1/2 # observed + # >observed)
0.216
0.168
p (>= observed)
0.240
0.185
p (# >observed)
0.140 *
0.098 **
p (1/2 # observed + # >observed)
0.150 *
0.106 *
p (>= observed)
0.159
0.113 *
***: significant at 5% level; **: significant at 10% level; * significant at 15% level
The simulation result moderately supports the claim that the hypothesized link A is stronger than
expected by chance. The p-values for completeness are around 0.10 under both random sampling schemes.
It seems the link is not significantly more accurate than expected by chance. The simulated p-values are
around 0.20.
The observed completeness C = 24.1%, and accuracy A = 44.2%, and the observed S =
4.67E-3, compared with the theoretical maximum for S is 0.014. Moreover P(M) and q(N) can be estimated
from the simulation and hence we can estimate the three local minimum under limiting circumstances 2, 3
and 4 discussed in the end of section 2. Table 2 has the comparison on S values under both randomization
and limiting circumstances. The observed S is above all the estimated local minimums representing the
strength of a random link.
Table 2. Estimated limiting extrema of combined measure S
Limiting case defined in section 2
Conditional
Unconditional
2 (local minimum)
4.06E-3
3.89E-3
3 (local minimum)
1.69E-3
1.60E-3
4 (local minimum)
2.37E-3
2.29E-3
1 (Theoretical maximum)
1.44E-2
Observed
4.67E-3
The count of non-empty QTLs and covered genes can be used to construct a chi-square type of test.
The test statistic is defined as
T
2
i 1
( X i - EX i ) 2
~ T21 under H 0 : the link is no different from random,
EX i
(7)
X i ni , mi
where EXi under H0 can be estimated by random sampling genes. The result p-values are in table 3.
Table 3. p-value from the chi-square test for the hypothesis H0: the link is not different from expected by chance.
Conditional
Unconditional
ni (Completeness)
<.001 ***
0.120 *
mi (Accuracy)
0.097 **
0.245
***: significant at 5% level; **: significant at 10% level; * significant at 15% level
A third test approach is based on the risky assumption that chromosomes are random samples from the
same population when measuring the strength of a link. Then the three measures we used can be seen as
random samples from two populations: one for the hypothesized link between QTL and NA genes, the other
for the random link representing background strength. The measures are paired on each chromosome. A
paired two-sample t-test or Wilcoxon sign-rank test (Myles et al, 1999) can then be applied. The results are
in table 4.
Table 4. p-value from the paired t and Wilcoxon sigh-rank test for the hypothesis H0: the link is not stronger than expected by chance.
Test
C (Completeness)
Conditional
Unconditional
0.106 *
0.100 **
Wilcoxon
0.196
0.209
Paired t
0.199
0.316
Wilcoxon
0.261
0.290
Paired t
0.191
0.180
Wilcoxon
0.275
0.275
Paired t
A (Accuracy)
S (Combined)
***: significant at 5% level; **: significant at 10% level; * significant at 15% level
The data and codes in R can be accessed from http://www.stat.psu.edu/~hanbing/qtlpaper/.
4 Conclusion and discussion
The link shows more difference in terms of completeness under both randomization schemes.
Meanwhile the difference in accuracy is weaker. Using the simulated one-sided p-value in table 1 and the
chi-square test on count, we can conclude that NA genes are significantly more complete in QTL spans than
by chance at least 15% significance level. However, it seems that QTL is not quite accurate in terms of
matching NA genes, i.e. most tests fail to reject null hypothesis even at 15% level. The combined measure S
strikes a balance between completeness and accuracy. The simulated one-sided p-values still reject the null
hypothesis in most cases. The p-values from those paired tests including both t test and Wilcoxon test in
table 4 should be taken carefully. The assumption that chromosomes are i.i.d. sample from a population is
dubious. From figure 1 at least three faces of chromosomes distinct apparently among chromosomes: length,
number and location of NA genes, and number and location of QTLs. We noticed that the paired tests
produce p-values with similar patterns to other tests but larger values. Even though we still reject the null
hypothesis for completeness by paired t-test. In sum with moderate evidence it can be concluded that the
link between QTL and NA genes is stronger than by chance. Particularly QTLs cover NA genes more
completely than by chance, while there could exist redundant QTLs such that the accuracy is not very
significantly different from by chance.
Completeness, accuracy and the combine measure have been proposed as methods to determine
whether a set of QTLs and a set of genes are associated. The statistical significance of the association can
be estimated by selecting sets of genes at random from the population of genes from which the gene set was
determined. A strong association was expected between the NA genes and the drug abuse QTLs. However,
this association is only moderately stronger than expected by chance. A possible reason is that the randomly
selected genes were selected from those represented on the Affymetrix® array U74Av2 which consists of
about one third of the whole genome. A second possibility is that there are considerably many QTLs
without association to the NA genes that result in a worse accuracy.
References
Carelli RM, and Wightman RM (2004) Functional microcircuitry in the accumbens underlying drug
addiction: insights from realtime signaling during behavior, Curr Opin Neurobiol. 14, 763-768.
Fischer, G, Ibrahim, SM, Brockmann, GA, Pahnke, J, Bartocci, E, Thiesen, H, Serrano-Fernandez, P, and
Molle, S. (2003) Expressionview: visualization of quantitative trait loc and geneexpression data in
Ensembl. Genome Biology, 4: R77.
Gentleman, RC, Carey, VJ, Bates, DM, Bolstad, B, Dettling, M, Dudoit, S, Ellis, B, Gautier, L, Ge, Y, and
Gentry, J. (2004) Bioconductor: Open software development for computational biology and
bioinformatics. Genome Biology 5: R80.
Hollander, M., Wolfe DA. (1999) Nonparametric statistical inference 2nd ed. John Wiley & Sons. New
York, US.
Jung, M. (2003) unpublished honors BS thesis, The Pennsylvania State University.
Silver, LM. (1995) Mouse genetics: concepts and applications. Oxford University Press, Oxford, UK.
Spellman PT, Rubin GM. (2002) Evidence for large domains of similarly expressed genes in the
Drosophila genome. Journal of Biology 1:5.1-5.
Voigt C, Moller S, Ibrahim SM, Serrano-Fernandez P. (2004). Non-linear conversion between genetic and
physical chromosomal distances. Bioinformatics. 20:1966-1967.
Wang J, Williams RW, Manly KF. (2003) WebQTL: Web-based complex trait analysis. Neuroinformatics
1: 299-308.
Wayne, ML and Mclntyre, LM (2002) Combining mapping and arraying: an approach to candidate gene
identification. PNAS:Genetics, 99, 14903-14906.