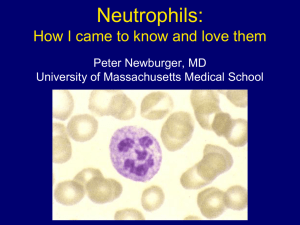
Table 2: Relative levels of cytokine gene expression in resting and
advertisement

Baines et al. 1 1 ONLINE DEPOSITORY 2 3 Differential gene expression and cytokine production from neutrophils in asthma 4 phenotypes 5 6 Katherine J Baines * PhD BBiomedSci(Hons) 1,2 7 Jodie L Simpson PhD BSc (Hons) 1,2, 8 Nikola A Bowden PhD BBiomedSci(Hons)3 9 Rodney J Scott PhD, DSc, FRCPath, FHGSA 3 10 Peter G Gibson MBBS FRACP 1,2, 11 1 12 Newcastle, Callaghan NSW 2308 AUSTRALIA 13 2 14 John Hunter Hospital, New Lambton NSW 2305 AUSTRALIA 15 3 16 Institute, The University of Newcastle, Callaghan NSW 2308 AUSTRALIA Priority Research Centre for Asthma and Respiratory Diseases, The University of Department of Respiratory and Sleep Medicine, Hunter Medical Research Institute Priority Research Center for Information Based Medicine, Hunter Medical Research 17 18 *Address for correspondence: Dr Katherine Baines 19 Level 3, HMRI, John Hunter Hospital 20 Locked Bag 1, Hunter Region Mail Centre 21 Newcastle, NSW 2310, AUSTRALIA 22 Email: katherine.baines@newcastle.edu.au 23 Phone: 61 2 49855766 Fax: 61 2 49855850 24 25 Baines et al. 26 2 Methods 27 28 Airway Neutrophil Isolation 29 CD16 positive cells were isolated from the remainder of the sputum sample using magnetic 30 cell separation. The cell pellet obtained was resuspended in phosphate-buffered saline (PBS) 31 supplemented with 0.1% bovine serum albumin (BSA) and 2mM EDTA (MACS buffer), and 32 incubated with CD16 microbeads (60uL per 50 x 106cells, Miltenyi Biotec, Gladbach, 33 Germany) for 30 minutes. The cells were applied to a LS MACS column surrounded by a 34 magnetic field (VarioMACS System, Miltenyi Biotec Gladbach, Germany), and the CD16- 35 cell fraction was eluted. The LS column was removed from the magnetic field and the CD16+ 36 neutrophil enriched cell fraction was eluted. Cytospins were prepared for each fraction, 37 stained (May-Grunwald Giemsa) and a differential cell count obtained from 400 cells, to 38 assess cell purity. Viability of the sputum neutrophil enriched fraction was assessed by trypan 39 blue and found to be 67%. 40 41 Peripheral Blood Neutrophil Isolation 42 Peripheral blood was collected in sodium citrate tubes. After centrifugation at 200g for 20 43 minutes the platelet rich plasma was removed. The remaining pellet was diluted and layered 44 over Percoll (GE Healthcare, Uppsala, Sweden) and centrifuged to deplete mononuclear cells. 45 After red blood cells were lysed, granulocytes were washed. The granulocyte cell pellet 46 obtained was resuspended in phosphate-buffered saline (PBS) supplemented with 0.1% 47 bovine serum albumin (BSA) and 2mM EDTA (MACS buffer), and incubated with CD16 48 microbeads (60uL per 50 x 106cells, Miltenyi Biotec, Gladbach, Germany). Neutrophils were 49 isolated from peripheral blood granulocytes via magnetic cell separation as per manufacturers 50 instructions (VarioMACS System, Miltenyi Biotec Gladbach, Germany). Cytospins were Baines et al. 3 51 prepared for each fraction, stained (May-Grunwald Giemsa) and a differential cell count 52 obtained from 400 cells, to assess cell purity. Neutrophil viability was assessed by trypan blue 53 exclusion and found to be 98%. 54 55 Neutrophil Culture and Stimulation 56 Highly pure blood neutrophils [100% (96-100%)] or the sputum neutrophil enriched cell 57 fraction [59% (30-78%) neutrophils; 35% (22-57%) macrophages] were resuspended in 58 phenol red free RPMI 1640 (Gibco Invitrogen, Mount Waverley, Australia) with 10mM 59 HEPES, 1% heat inactivated fetal calf serum and antibiotics (Penicillin/Streptomycin). The 60 cells were cultured at 1 x 105 (sputum) 1 x 106 (blood) cells/ml with or without 61 lipopolysaccharide (100ng/ml E.coli LPS, Sigma, Sydney, Australia) at 37˚C (5% CO2). Cell 62 cultures were collected at 24hrs. Neutrophil viability at 24 hours was assessed by trypan blue 63 exclusion and found to be 95% for resting neutrophils and 96% for LPS stimulated 64 neutrophils. Cell free supernatants were prepared and cell pellets were and stored in buffer 65 RLT (Qiagen, Hilden, Germany) at -80˚C. 66 67 68 69 70 71 72 73 74 75 Baines et al. 4 76 Results 77 Table E1: Clinical characteristics of participants with eosinophilic and non-eosinophilic 78 asthma selected for microarray analysis. 79 Eosinophilic Non-Eosinophilic Asthma Asthma n 5 4 Age years 63 (55-69) 70 (64-75) 0.33 Gender M | F 3|2 2|2 1.00 Atopy n (%) 4 (80) 3 (75) 1.00 FEV1 % predicted 66 (15) 53 (15) 0.21 FEV1/FVC % 80 (7) 71 (9) 0.15 ICS ‡‡ dose (μg) median (IQR) 1000 (500-2000) 2000 (1200-2500) 0.30 ‡‡ p ICS dose is calculated 1μg of beclomethasone = 1μg of budesonide = 0.5μg of fluticasone. Baines et al. 5 80 81 Table E2: Relative levels of cytokine gene expression in resting and LPS stimulated peripheral blood neutrophils isolated from participants with 82 eosinophilic (n=8) and non-eosinophilic asthma (n=9) and healthy controls (n=11). Resting 83 84 85 LPS Stimulated Healthy Controls Eosinophilic Asthma Non-Eosinophilic Asthma p Healthy Controls Eosinophilic Asthma Non-Eosinophilic Asthma p IL-8 2.24 (1.59-3.57) 1.07 (0.71-3.63) 3.07 (2.4-3.66) 0.16 54.2 (40.6-95.3) 26.4 (10.9-56.4) 44.6 (29.7-84.4) 0.23 IL-1β 0.07 (0.03-0.19) 0.01 (0.01-0.04) 0.03 (0.02-0.04) 0.07 1.4 (0.6-2.0) 0.4 (0.1-0.75)* 0.7 (0.2-1.3) 0.03 TNF-α 0.02 (0.01-0.06) 0.02 (0.01-0.03) 0.08 (0.02-0.1) 0.23 1.2 (0.6-1.7) 1.5 (0.4-1.9) 1.0 (0.6-1.5) 0.83 OSM 0.18 (0.07-0.24) 0.1 (0.07-0.15) 0.15 (0.13-0.18) 0.29 0.66 (0.35-0.84) 0.64 (0.26-1.09) 0.56 (0.46-0.72) 0.87 TLR2 0.6 (0.3-0.9) 0.14 (0.06-0.4)* 0.2 (0.2-0.3) 0.02 3.9 (2.5-8.4) 2.1 (1.6-3.4) 2.3 (1.1-2.9)* 0.02 TLR4 0.5 (0.2-0.8) 0.2 (0.2-0.5) 0.4 (0.3-0.4) 0.48 0.9 (0.5-1.0) 1.0 (0.5-1.3) 0.6 (0.5-1.0) 0.66 *p<0.008 versus healthy controls as determined by Kruskal-Wallis non parametric test for significance Baines et al. 6 86 87 Table E3: Mean fold changes in gene expression of selected targets identified to be 88 differentially expressed by microarrays and confirmed by relative real-time PCR. Gene Symbol Array Fold Change Real-Time PCR Fold Change PCR t test p value Altered by LPS stimulation (n=9) TLR2 + 11.7 + 20.6 <0.01 OSM + 3.2 + 14.3 <0.01 IL-8 + 9.2 + 23.2 <0.01 Altered in resting neutrophils in NEA (n=4) versus EA (n=5) GADD45B +3.1 +1.6 0.01 TNFRSF14 +2.2 +1.6 0.05 Altered in LPS stimulated neutrophils in NEA (n=4) versus EA (n=5) 89 90 91 92 93 94 95 96 97 98 PLAU +3.4 +1.1 0.55 CCL23 -5.9 -7.1 0.01 Baines et al. 99 100 7 Table E4: Spearman Rank Correlation Analysis between the expression of evaluated upregulated genes in noneosinophilic asthma and lung function (FEV1% predicted; n=9). FEV1% predicted GADD45B IRAK3 HM74 GADD45B IRAK3 HM74 MAIL PI3 STX4A HLAE HDAC5 TNFRSF14 -0.70b -0.77b -0.72b -0.78b -0.83c -0.67c -0.67b -0.77a -0.60a 0.68b 0.77b 0.63a 0.72b 0.80c 0.73b 0.95c 0.92c 0.82c 0.97c 0.75b 0.80c 0.70b 0.53 0.60a 0.77b 0.70b 0.80c 0.92c 0.63a 0.83c 0.80c 0.75b 0.65a 0.45 0.60a 0.72b 0.53 0.62a 0.70b 0.77b 0.78b 0.72b 0.63a 0.77b MAIL PI3 STX4A HLAE HDAC5 101 102 103 Data shows the Spearman rank correlation coefficient (r). p<0.10, b p<0.05 c p<0.01 a 0.80c