بالإشارة إلى الموضوع أعلاه،
advertisement

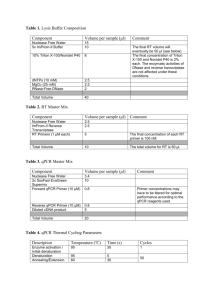
Administrative Information المعلومات االدارية Project Title - عنوان المشروع Development of a Rapid Test for Monitoring the Microbiological Quality of Marine Recreational Waters in Lebanon Principal Investigator - الباحث الرئيسي رقم الهاتف Telephone العنوان االلكتروني e-mail 70-164389 ps00@aub.edu.lb العنوان Address الوظيفية Post المؤسسة Institution االسم Name Assistant Professor American University of Beirut Pascal E. Saikaly Co-Workers - الباحثون المشاركون العنوان االلكتروني e-mail mfadel@aub.edu.lb eb01@aub.edu.lb gayoub@aub.edu.lb 1 year المؤسسة Institution American University of Beirut American University of Beirut American University of Beirut االسم Name Mutasem ElFadel Elie K. Barbour George M. Ayoub : Duration -المدة التعاقدية للمشروع Scientific Information العلمية المعلومات ّ Objectives - الهدف The objective of the study was to develop a novel DNA-based molecular test that allows for the rapid monitoring (less than 4h) of the microbiological quality of recreational waters in Lebanon. Achievements -أالنجازات المحققة 1. Construction of a blue LED light apparatus. The constructed light apparatus consists of 21 blue LEDS, a Fresnel lens and a convex lens. The blue LEDs are collectively used as a light source and the two lenses are used to focus the light on the PMA-treated Enterococcus faecalis cells which are collected on polycarbonate filters. The blue LED light apparatus is used for analyzing large-volume marine water samples. 2. Optimization of cell recovery from polycarbonate filters. The ability of different eluents to recover cells from polycarbonate filters was tested. The tested eluents were PBS (1X), STE and Tween 80 (0.1%). STE achieved the highest percent recovery (97%) of Enterococcus faecalis cells from polycarbonate filters. 3. Optimization of DNA extraction. The ability of different chemical and physical lysis methods to extract DNA from environmental samples was tested in this study. Of the different methods tested, hot detergent treatment was selected to extract genomic DNA because it was the most effective method in lysing cells. 4. Optimization of PMA treatment in phosphate buffered saline (PBS). The PMA protocol for the differentiation of viable and dead E. faecalis cells was optimized in PBS. The maximum differentiation achieved between viable and dead E. faecalis cells was 12 Ct cycles for an incubation period of 5 minutes, a light exposure period of 30 minutes and a PMA concentration of 15 µM. 5. Optimization of PMA treatment in seawater. The PMA protocol for the differentiation of viable and dead E. faecalis cells was optimized in seawater. The maximum differentiation achieved between viable and dead E. faecalis cells was 9 Ct cycles for an incubation period of 20 minutes, a light exposure period of 30 minutes and a PMA concentration of 50 µM. 6. Construction of a qPCR standard curve. A qPCR standard curve was generated for the quantification of total E. faecalis cells in seawater samples. Quantitative amplification parameters for E. faecalis were optimal with linearity over a 6-log dynamic range (R2 = 0.983) and an overall PCR amplification efficiency of 0.93. 7. Construction of a PMA-qPCR standard curve. A PMA-qPCR standard curve was generated for the quantification of viable E. faecalis cells in seawater samples. Quantitative amplification parameters for E. faecalis were optimal with linearity over a 6-log dynamic range (R2 = 0.953) and an overall amplification efficiency of 1.04. Publications & Communications - المنشورات والمساهمات في المؤتمرات A manuscript is under preparation to be submitted to a high-impact peer-reviewed journal article in the field of environmental science and engineering. Abstract - موجز عن نتائج البحث Quantitative polymerase chain reaction (qPCR) assays were previously developed for the detection and quantification of the fecal indicator bacteria, Enterococcus spp., in marine recreational waters as an alternative to the time-consuming standard membrane filtration (MF) method. The developed qPCR assays, however, cannot differentiate between viable and dead Enterococcus cells. Recently, propidium monoazide (PMA), a DNA binding dye, has been successfully integrated into qPCR assays for the differentiation of viable and dead bacterial cells. In the present study a PMA-qPCR assay was developed for the detection and quantification of viable Enterococcus spp. in marine recreational waters. The microorganism Enterococcus faecalis was used in this study as a representative organism of Enterococcus spp. The PMA treatment for the differentiation of viable and dead E. faecalis cells was optimized in phosphate buffered saline (PBS) and seawater. The optimized PMA treatments were successful, in significantly reducing the qPCR signal of dead E. faecalis cells without affecting the qPCR signal of viable E. faecalis cells. The optimized PMA treatment for PBS samples reduced the qPCR signal of dead E. faecalis cells by 4 log10 units while the optimized PMA treatment for marine water samples reduced the qPCR signal of dead E. faecalis cells by only 3 log10 units. A PMA-qPCR standard curve for the quantification of viable Enterococcus spp. was generated using seawater samples spiked with serially diluted concentrations of E. faecalis cells. The generated PMA-qPCR standard curve was linear over a 6-log dynamic range and had a detection limit of 100 cells per mL. In conclusion, the developed PMA-qPCR assay could provide a rapid and accurate method for quantifying viable Enterococcus spp. in marine recreation waters.