file - BioMed Central
advertisement

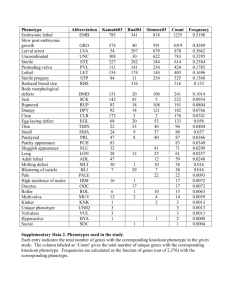
Additional material for Interlinked nonlinear sub-networks underlie the formation of cellular patterns in Arabidopsis epidermis: a spatial model of coupled gene regulatory networks. Mariana Benítez, Carlos Espinosa-Soto1, Pablo Padilla-Longoria and Elena R. AlvarezBuylla1 Logical rules constitute the precise description of the network topology and of the function with which the nodes’ values are calculated in every time-step: gn(t+1)=Fn(gn1(t),gn2(t),..., gnk(t)). Values of the regulators are on the left side of the tables, while the regulated node or nodes are on the right side. X indicates that, given the entrances of the rest of the table, the node can take any value. Y and Z are temporal variables that integrate the values of two groups of nodes (WER/GL1, GL3, EGL3, TTG1 and TRY, CPC, ETC, respectively) and are then used to estimate the expression value of other nodes. The definition of Y and Z facilitates the presentation and implementation of the logical rules, but does not affect the synchronic updating schema or the interactions among elements of the network. 1 eabuylla@gmail.com 1 1. Logical rules for updating the discrete gene regulatory network model for the leaf epidermis. GL1 0 X X 1 2 2 Inputs Output GL3 EGL3 TTG1 Y X X GL3+EGL3=0 GL3+EGL3<3 2 2 2 X 1 2 ELSE Inputs CPC TRY 0 0 2 1 Inputs Y Z 0 0 Y<Z 1 2 1 2 0 0 1 1,2 GL1 1 1 2 2 2 2 X X 0 1,2 1,2 1,2 ETC CPC+ETC<2 2 0 CPC+ETC>0 2 X ELSE 0 0 0 2 2 2 1 Output Z 0 0 2 2 1 Outputs GL3 EGL3 GL2 TTG1 ETC CPC 0 0 1 1 1 1 0 0 1 1 1 1 0 0 1 2 1 2 Input Output GL2 TRY 0 1 2 0 1 2 2 1 1 1 1 1 1 0 0 1 2 1 2 0 0 1 2 1 2 2. Logical rules for updating the discrete gene regulatory network model for the root epidermis. Inputs Output GL3 EGL3 TTG1 Y WER 0 X X 1,2 2 2 X X GL3+EGL3<3 0 0 2 2 1 2 2 0,1 ELSE Inputs TRY CPC 0 X 2 Inputs Y Z 0 0 0 1 1 1 2 2 2 0 1 2 0 1 2 0 1 2 ETC 0,1 2 X ELSE 0 X X 0 0 0 2 2 2 1 Output Z 0 2 2 1 Outputs CPC WER TTG1 GL3 EGL3 1 2 2 0 1 1 0 0 2 X 0 1 X 1 1 1 2 2 0 1 1 0 0 2 0 0 0 1 0 0 2 2 1 1 0 0 1 0 0 2 2 1 Input Output GL2 TRY 0 1 2 0 1 2 3 1 1 1 1 1 1 1 1 1 GL2 ETC 0 0 0 1 0 0 2 2 1 0 0 0 1 0 0 1 1 1 TABLE S1. Reported and simulated phenotypes corresponding to single and double loss of function (italics low case), as well as to overexpression (+) lines for the leaf metaGRN. All simulations were carried out with the same parameter values (DCPC= 0.05, DTRR= 0.05, DTTG= 0.03). Cell type patterns in the leaf epidermis of Arabidopsis thaliana Genotype Reported phenotype Simulated phenotype Wild type Trichomes spaced out with no clusters More trichomes than in wt, no clusters High cluster probability No phenotype Enhancement of the cpc phenotype High number of trichomes and clusters High cluster probability (try-like phenotype) No trichomes No trichomes More trichomes than in wt Wt phenotype ttg phenotype No trichomes Many more trichomes than in wt, but still with few pavemente cells Trichomes spaced out with no clusters ~5% more tricomes than in wt, no clusters High cluster probability No phenotype Enhancement of the cpc phenotype with few clusters High number of trichomes and clusters High cluster probability (try-like phenotype) No trichomes No trichomes More trichomes than in wt Wt phenotype ttg phenotype No trichomes Only trichomes and no pavement cells cpc try etc cpc etc cpc etc try cpc try ttg bhlh bHLH (+) ttg bHLH (+) ttg GL1(+) gl1 GL1 (+) bHLH (+) 4 TABLE S2. Reported and simulated phenotypes corresponding to single and double loss of function (italics low case), as well as to overexpression (+) lines for the root metaGRN. All simulations were carried out with the same parameter values (DCPC= 0.01, DGL3= 0.01, DEGL3= 0.01). Cell type patterns in the root epidermis of Arabidopsis thaliana Genotype Wild type scm wer bhlh cpc try cpc etc CPC (+) WER (+) Reported phenotype Simulated phenotype Alternated bands of hair and Alternated bands of hair and no-hair cells no-hair cells Hairs and no-hair cells Hairs and no-hair cells without banded pattern. without banded pattern. Many ectopic hairs Only hair cells Remarkable increase in the Remarkable increase in the number of hair cells number of hair cells Ectopic atrichoblasts Ectopic atrichoblasts Ectopic atrichoblasts Ectopic atrichoblasts Enhanced cpc phenotype Enhanced cpc phenotype Ectopic hair cells Ectopic hair cells Wt phenotype Wt-like phenotype with some ectopic cells 5