Supplementary material - Springer Static Content Server
advertisement

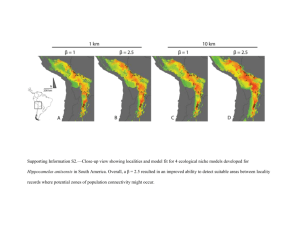
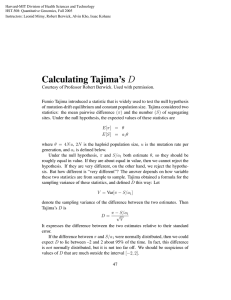
ELECTRONIC SUPPLEMENTARY MATERIAL Matrilineal evidence for demographic expansion, low diversity and lack of phylogeographic structure in the Atlantic forest endemic Greenish Schiffornis Schiffornis virescens (Aves: Tityridae) Journal of Ornithology Cabanne G Sa,b,c, Sari E Rd, Meyer Da, Santos F Rd, Miyaki CYa aDepartamento de Genética e Biologia Evolutiva, Instituto de Biociências, Universidade de São Paulo, Rua do Matão 277, 05508–090, São Paulo, SP, Brazil. bCONICET, c División Av. Rivadavia 1917, Ciudad de Buenos Aires (C1033AAJ), Argentina. de Ornitologia, Museo Argentino de Ciencias Naturales “B. Rivadavia”, Ángel Gallardo 470, Ciudad de Buenos Aires (C1405DJR), Argentina. dDepartamento de Biologia Geral, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Av. Antônio Carlos 6627, 31270-901, Belo Horizonte, MG, Brazil. Supplementary material Material and Methods Coalescent simulations In order to evaluate different demographic histories we simulated possible demographic scenarios in the program BAYESSC, a modification of the software SERIAL SIMCOAL (Anderson et al. 2005; Chan et al. 2006), and evaluated the goodness of fit of the observed data to the simulations. The tested models (detailed in Figure 2) differed in the timing, number, and intensity of bottlenecks, and also in the number of populations. The populations in model G were originated before the Pleistocene, as suggested by the divergence of S. virescens with its sister species S. turdina (See Results) In the simulations we organized the sampling localities (Table 1) by proximity into five populations (pop); pop 1: localities 1-5; pop 2: locality 6-8, 11; pop 3: localities 12-14; pop 4: localities 9, 10, 15-17; pop 5: localities 18 and 19. To model panmixia we collapsed the five populations into one at the first generation. Bottlenecks reduced Ne (effective size of genes) to 1-10% and 10-100% of the present size. We obtained the species Ne from theta (Θ=2μNe) estimated in LAMARC 2.1.2b (Kuhner 2006). Each population in the islands models, as well as the ancestral population of the model G, presented an effective size equal to 1/5 of the species Ne. Estimations of Θ used the F84 model of sequence evolution, empirical base frequencies and transition/transversion ratios, with a Markov chain Monte Carlo with default setting. Effective number of genes was introduced in BAYESSC infiles as a normal distribution with mean equal to the maximum likelihood estimation of Ne and a standard deviation estimated from Θ confidence interval. For the CR substitution rate we used 1.67 x 10-8 substitution/site/MY (SE 1.67 x 10-9). We had previously obtained this value in phylogenetic analyses in BEAST which used cytb and CR sequences and estimated the CR rate in relation to a cytb rate (2.1 % divergence/MY, Weir and Schluter 2008). The transition bias was 0.79, the mutation rate gamma distribution was 0.7576, and the number of mutation categories was six. We assumed a generation time of one year. For each model we ran 1,000 simulations. Then, for each simulated data we estimated summary statistics for the complete data set to obtain null distributions against which we tested the observed data. Summary statistics were: segregating sites, nucleotide diversity, Tajima’s D (Tajima 1989), Fu’s Fs (Fu 1997), and Φst (Excoffier et al. 1992). For evaluating the goodness of fit of the observed data to simulated data 1) we used the two-tailed empirical likelihood pi of each summary statistics i; pi 1 2 10.5 p (eq.1), being p the proportion of simulated values equal or higher than the observed summary statistic. Then, 2) we combined the five pi values by obtaining Cobs 2i1 ln pi (eq.2) and getting its significance. The significance was assessed by comparing k Cobs against a null distribution of C obtained according to Voight (2005) and Fabre (2009). Briefly, for each simulated dataset, each value of summary statistic was compared with the other values representing the empirical distribution of the statistic from simulation. Specifically, we treated the value of each summary statistic as the observed value and calculated with eq. 1 its psim-value relative to the remaining 999 simulated data. Then, the null distribution of C was obtained by combining with eq. 2 values of psim across summary statistics and the significance of Cobs was obtained as in step 1 (eq.1). This procedure generated two-tailed global p-values associated to each model that we used to evaluate plausibility of models. REFERENCES Anderson CNK, Ramakrishnan U, Chan YL, Hadly EA (2005) Serial SimCoal: A population genetics model for data from multiple populations and points in time. Bioinformatics 21 (8):1733-1734. doi:10.1093/bioinformatics/bti154 Chan YL, Anderson CNK, Hadly EA (2006) Bayesian estimation of the timing and severity of a population bottleneck from ancient DNA. Plos Genetics 2 (4):451460. doi:e59 10.1371/journal.pgen.0020059 Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes - application to human mitochondrial-DNA restriction data. Genetics 131 (2):479-491 Fabre V, Condemi S, Degioanni A (2009) Genetic Evidence of Geographical Groups among Neanderthals. PLoS ONE 4 (4):e5151 Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147 (2):915-925 Kuhner MK (2006) LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics 22 (6):768-770 Tajima F (1989) Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 (3):585-595 Voight BF, Adams AM, Frisse LA, Qian Y, Hudson RR, Di Rienzo A (2005) Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci U S A 102 (51):18508-18513. doi:10.1073/pnas.0507325102 Weir JT, Schluter D (2008) Calibrating the avian molecular clock. Mol Ecol 17 (10):2321-2328