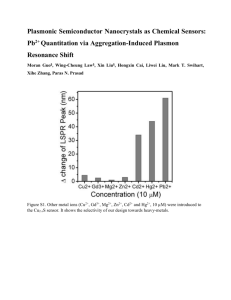
Supplementary Figure Supplementary Figure 1. Chloroplast
advertisement

Supplementary Figure Supplementary Figure 1. Chloroplast haplotype diversity for 27 woody plant species including Rhododendron dealavayi var. delavayi. Tree species found in south-west China are shown in blue bars. References 1 Chen, S.-C.; Zhang, L.; Zeng, J.; Shi, F.; Yang, H.; Mao, Y.-R.; Fu, C.-X., Geographic variation of chloroplast DNA in Platycarya strobilacea (Juglandaceae). Journal of Systematics and Evolution 2012, 50, (4), 374-385. 2 Chiang, T. Y.; Chiang, Y. C.; Chen, Y. J.; Chou, C. H.; Havanond, S.; Hong, T. N.; Huang, S., Phylogeography of Kandelia candel in East Asiatic mangroves based on nucleotide variation of chloroplast and mitochondrial DNAs. Molecular Ecology 2001, 10, (11), 2697-2710.3 3 Jia, D.-R.; Abbott, R. J.; Liu, T.-L.; Mao, K.-S.; Bartish, I. V.; Liu, J.-Q., Out of the Qinghai– Tibet Plateau: evidence for the origin and dispersal of Eurasian temperate plants from a phylogeographic study of Hippophaë rhamnoides (Elaeagnaceae). New Phytologist 2012, 194, (4), 1123-1133. 4 Huang, C.; Hung, K.; Hwang, C.; Huang, J.; Lin, H.; Wang, W.; Wu, P.; Hsu, T.; Chiang, T., Genetic population structure of the alpine species Rhododendron pseudochrysanthum sensu lato (Ericaceae) inferred from chloroplast and nuclear DNA. BMC Evolutionary Biology 2011, 11, (1), 108. 5 Huang, C.; Hung, K.; Hwang, C.; Huang, J.; Lin, H.; Wang, W.; Wu, P.; Hsu, T.; Chiang, T., Genetic population structure of the alpine species Rhododendron pseudochrysanthum sensu lato (Ericaceae) inferred from chloroplast and nuclear DNA. BMC Evolutionary Biology 2011, 11, (1), 108. 6 Liu, Y.; Yang, S.-x.; Ji, P.-z.; Gao, L.-z., Phylogeography of Camellia taliensis (Theaceae) inferred from chloroplast and nuclear DNA: insights into evolutionary history and conservation. BMC Evolutionary Biology 2012, 12, (1), 92. 7 Wang, J.; Gao, P.; Kang, M.; Lowe, A. J.; Huang, H., Refugia within refugia: the case study of a canopy tree (Eurycorymbus cavaleriei) in subtropical China. Journal of Biogeography 2009, 36, (11), 2156-2164. 8 Okaura, T.; Quang, N. D.; Ubukata, M.; Harada, K., Phylogeographic structure and late Quaternary population history of the Japanese oak Quercus mongolica var. crispula and related species revealed by chloroplast DNA variation. Genes & Genetic Systems 2007, 82, (6), 465-477 9 Gao, L. M.; MÖLler, M.; Zhang, X. M.; Hollingsworth, M. L.; Liu, J.; Mill, R. R.; Gibby, M.; Li, D.Z., High variation and strong phylogeographic pattern among cpDNA haplotypes in Taxus wallichiana (Taxaceae) in China and North Vietnam. Molecular Ecology 2007, 16, (22), 4684-4698. 10 Xu, T.; Abbott, R.; Milne, R.; Mao, K.; Du, F.; Wu, G.; Ciren, Z.; Miehe, G.; Liu, J., Phylogeography and allopatric divergence of cypress species (Cupressus L.) in the Qinghai-Tibetan Plateau and adjacent regions. BMC Evolutionary Biology 2010, 10, (1), 194. 11 Zhang, T.-C.; Comes, H. P.; Sun, H., Chloroplast phylogeography of Terminalia franchetii (Combretaceae) from the eastern Sino-Himalayan region and its correlation with historical river capture events. Molecular Phylogenetics and Evolution 2011, 60, (1), 1-12. 12 Li, Y.; Yan, H.-F.; Ge, X.-J., Phylogeographic analysis and environmental niche modeling of widespread shrub Rhododendron simsii in China reveals multiple glacial refugia during the last glacial maximum. Journal of Systematics and Evolution 2012, 50, (4), 362-373. 13 Liu, J.; Sun, P.; Zheng, X.; Potter, D.; Li, K.; Hu, C.; Teng, Y., Genetic structure and phylogeography of Pyrus pashia L. (Rosaceae) in Yunnan Province, China, revealed by chloroplast DNA analyses. Tree Genetics & Genomes 2013, 9, (2), 433-441. 14 Huang, S. S. F.; Hwang, S.-Y.; Lin, T.-P., Spatial pattern of chloroplast DNA variation of Cyclobalanopsis glauca in Taiwan and East Asia. Molecular Ecology 2002, 11, (11), 2349-2358. 15 Lee, J.-H.; Lee, D.-H.; Choi, B.-H., Phylogeography and genetic diversity of East Asian Neolitsea sericea (Lauraceae) based on variations in chloroplast DNA sequences. J Plant Res 2013, 126, (2), 193-202. 16 Li, X.-H.; Shao, J.-W.; Lu, C.; Zhang, X.-P.; Qiu, Y.-X., Chloroplast phylogeography of a temperate tree Pteroceltis tatarinowii (Ulmaceae) in China. Journal of Systematics and Evolution 2012, 50, (4), 325-333 17 17 Kyoda, S.; Setoguchi, H., Phylogeography of Cycas revoluta Thunb. (Cycadaceae) on the Ryukyu Islands: very low genetic diversity and geographical structure. Plant Syst Evol 2010, 288, (3-4), 177-189. 18 This study (Rhododendron dealavayi var. delavayi) 19 Zou, J.-B.; Peng, X.-L.; Li, L.; Liu, J.-Q.; Miehe, G.; Opgenoorth, L., Molecular phylogeography and evolutionary history of Picea likiangensis in the Qinghai–Tibetan Plateau inferred from mitochondrial and chloroplast DNA sequence variation. Journal of Systematics and Evolution 2012, 50, (4), 341-350. 20 Huang, S.-F.; Hwang, S.-Y.; Wang, J.-C.; Lin, T.-P., Phylogeography of Trochodendron aralioides (Trochodendraceae) in Taiwan and its adjacent areas. Journal of Biogeography 2004, 31, (8), 1251-1259 21 Zhang, Q.; Chiang, T. Y.; George, M.; Liu, J. Q.; Abbott, R. J., Phylogeography of the Qinghai-Tibetan Plateau endemic Juniperus przewalskii (Cupressaceae) inferred from chloroplast DNA sequence variation. Molecular Ecology 2005, 14, (11), 3513-3524. 22 Meng, L.; Yang, R. U. I.; Abbott, R. J.; Miehe, G.; Hu, T.; Liu, J., Mitochondrial and chloroplast phylogeography of Picea crassifolia Kom. (Pinaceae) in the QinghaiTibetan Plateau and adjacent highlands. Molecular Ecology 2007, 16, (19), 4128-4137. 23 Wang, J.; Wu, Y.; Ren, G.; Guo, Q.; Liu, J.; Lascoux, M., Genetic Differentiation and Delimitation between Ecologically Diverged Populus euphratica and Populus pruinosa. PLoS ONE 2011, 6, (10), e26530. 24 Wang, J.; Wu, Y.; Ren, G.; Guo, Q.; Liu, J.; Lascoux, M., Genetic Differentiation and Delimitation between Ecologically Diverged Populus euphratica and Populus pruinosa. PLoS ONE 2011, 6, (10), e26530. 25 Jiang, Z. Y.; Peng, Y. L.; Hu, X. X.; Zhou, Y. F.; Liu, J. Q., Cytoplasmic DNA variation in and genetic delimitation of Abies nephrolepis and Abies holophylla in northeastern China. Canadian Journal of Forest Research 2011, 41, (7), 1555-1561. 26 Mathiasen, P.; Premoli, A. C., Out in the cold: genetic variation of Nothofagus pumilio (Nothofagaceae) provides evidence for latitudinally distinct evolutionary histories in austral South America. Molecular Ecology 2010, 19, (2), 371-385. 27 Jiang, Z. Y.; Peng, Y. L.; Hu, X. X.; Zhou, Y. F.; Liu, J. Q., Cytoplasmic DNA variation in and genetic delimitation of Abies nephrolepis and Abies holophylla in northeastern China. Canadian Journal of Forest Research 2011, 41, (7), 1555-1561.