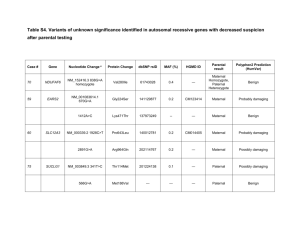
Supplementary Figure 1: Ingenuity Pathways Analysis of mutated

Supplementary Figure 1: Ingenuity Pathways Analysis of mutated genes in BPDCN. (A)
Substantial enrichment of pathways involved in dermatological diseases and conditions, genetic disorder, hematological disease, among others; (B) Analysis of the ten validated genes. Although the low number of genes resulted in non-statically significant values, pathways such as spliceosomal cycle appeared in as enriched pathways.
Supplementary Figure 2: Sanger sequencing chromatograms of IKZF3 and ZEB2 mutations (TM: tumoral; NT: non- tumoral)
Supplementary Figure 3: Pairwise co-occurrence of (A) TET1-2 and (B) IDH1-2 mutations in
BPDCN. A circos diagram depicts the relative frequency and pairwise co-occurrence of other mutations with TET1-2 or IDH1-2 mutations in the entire cohort. The length of the arc indicates the frequency of mutations in the first gene, and the width of the ribbon represents the percentage of
BPDCN patients with TET1-2 or IDH1-2 mutations who bear the second gene mutation. Pairwise cooccurrence of mutations is denoted only once, beginning with the TET1-2 or IDH1-2 gene in the clockwise direction.
Supplementary Figure 4: Target NGS result by pathways. (A) A Circos diagram depicts the relative frequency and pairwise co-occurrence of mutations by pathways in the BPDCN patients. The length of the arc corresponds to the frequency of mutations in the first pathway, and the width of the ribbon corresponds to the percentage of patients who also had a mutation in the second pathway.
Pairwise co-occurrence of mutations is denoted only once, beginning with the first pathway in the clockwise direction; (B) Distribution of numbers and categories of unknown and pathogenic mutations among the 25 BPDCN cases. We do not identify any unknown nor pathogenic mutations among case 12 and 29 in the 38 studied genes, however, in case 8 we found mutations in almost all the represented pathways.
Supplementary Figure 5: Kaplan-Meier overall survival curves for ETV6 (p=0,022), TP53
(p=0,003), IKZF1/2/3 (p=0,073) and RAS (p=0,015). Patients with mutations in these genes had a reduced cumulative survival. As the number in each subgroup of patients with individual gene mutations was very small, we combined the subgroups based on the functional classes for the outcome analysis.
Supplementary Table 1: Main clinical characteristics of BPDCN patients.
Supplementary Table 2: 85 SNVs in 81 affected genes revealed by WES: 28 SNVs in case 1, 8 in case 2, and 49 in case
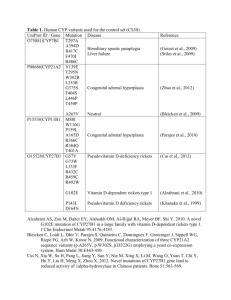
Supplementary Table 3: Genes included in the target next generation sequencing
N= Gene Coverage
1 ASXL1 100.0% CCDS13201.1
2 ATRX
3 CBL
4 CBLB
5 CBLC
100.0%
100.0%
100.0%
100.0%
CCDS14435.1, CCDS14434.1
CCDS8418.1
CCDS2948.1
Source
CCDS12643.1, CCDS46109.1
6 CEBPA 100.0% CCDS54243.1
7 DNMT3A 100.0% CCDS1718.2, CCDS33157.1, CCDS46232.1
8 ETV6
9 EZH2
100.0%
96.4%
CCDS8643.1
CCDS5892.1, CCDS5891.1
10 FLT3 100.0% CCDS31953.1
11 HOXA1 100.0% CCDS5401.1, CCDS5402.2
12 HOXB9 100.0% CCDS11534.1
13 IDH1 100.0% CCDS2381.1
14 IDH2
15 IKZF1
16 IKZF2
17 IKZF3
100.0%
100.0%
100.0%
100.0%
CCDS10359.1
NM_001220773, NM_001220767
CCDS2395.1, CCDS46507.1
CCDS11346.1, CCDS11351.1, CCDS11349.1
18 JAK2 100.0% CCDS6457.1
19 KDM6A 100.0% CCDS14265.1
20 KIT
21 KRAS
100.0%
100.0%
CCDS3496.1, CCDS47058.1
CCDS8702.1, CCDS8703.1
22 LUC7L2 100.0% CCDS43656.1
23 MPL 100.0% CCDS483.1
24 NBL1
25 NPM1
100.0%
100.0%
CCDS41278.1, CCDS196.1
CCDS4376.1, CCDS43399.1, CCDS4377.1
26 NRAS 100.0% CCDS877.1
27 PDGFRA 100.0% CCDS3495.1
28 RUNX1
29 SF3B1
99.9%
99.9%
NM_001001890, NM_001754, NM_001122607
CCDS33356.1, CCDS46479.1
30 SRSF2
31 TET1
32 TET2
33 TP53
34 U2AF1
35 UBE2G2
36 WT1
37 ZEB2
38 ZRSR2
100.0%
100.0%
100.0%
100.0%
100.0%
98.2%
100.0%
100.0%
97.5%
CCDS11749.1
CCDS7281.1
CCDS47120.1, CCDS3666.1
CCDS11118.1, CCDS45605.1, CCDS45606.1
CCDS33574.1, CCDS13694.1, CCDS42948.1
CCDS33586.1, CCDS13714.1
CCDS44562.1, CCDS7878.2, CCDS44561.1
CCDS54403.1, CCDS2186.1
CCDS14172.1
Number of target region: 469
Total target region size: 86266 bp
Coverage: 99,9% (86137 bp)
T
TCTGC
T
T
G
T
G
T
TCGAC
A
A
G
T
T
T
T
ACGCTCACCAAT
G
T
TA
CT
A
C
TCTCATA
A
A
CG
CG
TC
C
T
AG
T
G
T
CT
A
T
A
CG
T
CG
G
T
T
A
T
TCTGC
TTAG
A
T
T
C
T
T
T
T
AAC
AAC
T
C
A
G
T
T
CG
C
A
G
A
C
CT
T
T
Variant
TA
GT
T
A
C
T
A
T
C
A
A
G
G
C
G
T
C
C
C
C
T
AT
AT
A
C
C
C
C
T
C
C
AATAATTTT
G
C
T
C
T
C
C
T
T
G
T
A
C
A
C
C
C
T
T
AC
C
T
A
G
C
C
C
C
C
C
C
CA
C
G
G
TG
A
A
T
TG
TG
C
T
G
C
G
A
C
C
C
Ref
T
G
TC
AG
TTGTG
T
CCT
G
170819930
170837544
170819930
115258747
115258748
115252204
115258747
115258747
36252864
198266476
198266476
198257842
74732959
74732959
70332921
106157044
106164901
106197378
106180838
106196345
106156566
106196415
106156566
106158403
106164832
106193892
7578536
7577551
TET2
TET2
TET2
TET2
TP53
TP53
44514876
44514769
44524456
U2AF1
U2AF1
U2AF1
46197270 UBE2G2
145156878 ZEB2
145156878 ZEB2
145161489
145157495
15827389
15834002
ZEB2
ZEB2
ZRSR2
ZRSR2
SRSF2
SRSF2
TET1
TET2
TET2
TET2
TET2
TET2
TET2
TET2
NPM1
NPM1
NPM1
NRAS
NRAS
NRAS
NRAS
NRAS
RUNX1
SF3B1
SF3B1
SF3B1
213921691
37922598
37922621
37922598
5054619
55593690
25378562
25398282
25368410
170837544
170819932
170819952
11992124
148525964
148525964
148513776
148523616
28602329
46700467
209113112
90631934
90631934
90631934
50450305
Position
31022978
31024844
31022898
31025066
31021157
31021162
31023135
31022550
Gene
ASXL1
ASXL1
ASXL1
ASXL1
ASXL1
ASXL1
ASXL1
ASXL1
31024021
76939409
76939409
76888705
76776372
76812991
ASXL1
ATRX
ATRX
ATRX
ATRX
ATRX
105377837
45296756
CBLB
CBLC
25457243 DNMT3A
25536822 DNMT3A
11992124 ETV6
11992124 ETV6
IKZF2
IKZF3
IKZF3
IKZF3
JAK2
KIT
KRAS
KRAS
KRAS
NPM1
NPM1
NPM1
ETV6
EZH2
EZH2
EZH2
EZH2
FLT3
HOXB9
IDH1
IDH2
IDH2
IDH2
IKZF1
45,29
64,81
8,95
41,88
36,9
30
39,33
37,59
29
39,23
6,02
33,83
5,83
17,76
23,8
9,66
5,26
7,23
45,29
19,52
16,32
45,1
56,34
36,54
53,87
27,58
38,65
62,63
9,02
30,36
25,27
28,68
27,98
23,91
18,2
36,05
59,74
17,42
6,01
25,11
26,61
26,49
10,3
4,64
10,59
30,32
43,02
33,71
100
19,16
26,2
19,44
16,22
41,4
29,92
12,33
42,94
19,42
14,87
45,51
5,54
54,59
27,19
5,32
5,71
70,59
5,49
5,17
16,07
8,54
36,4
52,94
28,66
18,47
Var Freq
45,57
21,43
42,51
21,43
41,2
42,9
50,95
49,66
Supplementary Table 4: Pathogenic variants found using target next generation sequencing
170
108
525
1466
626
850
300
689
1000
808
646
788
223
210
380
765
930
133
103
670
458
290
4091
616
182
129
193
46
994
572
1253
765
727
835
1000
491
77
666
340
188
1311
264
30
214
516
1561
932
906
233
3408
2718
515
2166
2516
1551
850
1290
36
37
372
1183
981
168
199
684
17
841
1202
1927
282
210
17
1966
2497
Coverage
1457
28
1583
392
716
613
997
149
Case_25
Case_13
Case_01
Case_03
Case_08
Case_28
Case_07
Case_07
Case_39
Case_13
Case_18
Case_18
Case_08
Case_13
Case_14
Case_05
Case_05
Case_11
Case_03
Case_13
Case_13
Case_14
Case_18
Case_06
Sample
Case_09
Case_11
Case_14
Case_16
Case_22
Case_22
Case_41
Case_01
Case_08
Case_10
Case_11
Case_28
Case_42
Case_42
Case_42
Case_24
Case_13
Case_14
Case_06
Case_04
Codon Change
TTG TG[T/A] GTC
AGG [G/T]GA GGC
GGA [G/A]AA AAA
GGA [G/A]AA AAA
GCT TT[G/C] GTT
CTC T[C/T]A GAG
ACT TT[T/G] GAG
CCC T[T/C]C TGC
AGC [C/T]GC TTG
GGG G[A/C]C ACC
CAC C[G/A]G TTG
TTA CA[C/G] TCC
GGG G[C/T]G TGC
GCT C[G/A]C TCT
GGT C[G/A]T CAT
ATC C[G/A]G AAC
ATC C[G/A]G AAC
ATC C[G/A]G AAC
GTG G[C/T]T GAC
GCC [G/A]AA GCC
ATT T[T/G]G ACA
CCC [A/T]GA AAC
TCA [G/A]CA AAG
GGT [G/T]GC GTA
CCT [G/A]GC TGT
CTT [-/TAG] GCT
Case_28
Case_03
Case_08
Case_16
Case_18
Case_41
Case_04
Case_08
Case_09
Case_07
Case_14
Case_28
AAA [C/T]TT GCT
AAA [C/T]TT GCT
GCA G[G/A]T GGT
GCA [G/C]GT GGT
TCA [G/A]CC AAG
GCA G[G/C]T GGT
GCA G[G/A]T GGT
Case_14
Case_08
Case_02
Case_06
Case_07
Case_07
Case_22
Case_22
Case_24
Case_39
GGT [T/C]GT GAA
CGC C[C/A]C CCG
CGC C[C/A]C CCG
TCA [C/T]GA GTA
GAC [C/T]AA CAT
CAG C[A/G]T AAG
GGT T[G/T]T TCA
TCT T[C/A]A CAC
Case_41
Case_02
Case_03
Case_08
Case_07
Case_01
TGG [G/T]AA GGA
CGG [C/T]GA AAA
AAC [A/C]AG ATG
ATG [G/A]GC GGC
Case_25 GCG G[A/G]A AAG
Case_41 GAG [-/TATGAG] ATG
Case_02
Case_01
Case_18
TGC T[C/T]T CGG
CTT G[A/T]T TAC
Case_37
Case_42
Case_01
Case_06
Case_22
GGG C[T/G]T GGA
GAT [C/T]GA GCT
AA Change
C387*
G679*
E447K
E447K
L1708F
S2365L
F2210L
F388S
R882C
D11A
R502Q
H279Q
A680V
R183H
R132H
R140Q
R140Q
R140Q
A97V
E318K
L224W
R586*
A146T
G13C
G179S
L158*
L158F
L158F
G12R
G12R
A146T
G12A
G12D
C1204R
P122H
P122H
R276*
Q670*
H1925R
C1310F
S1604*
E1255*
R1473*
K132Q
G244S
E124G
E159EYE
S34F
D63V
L420R
R169*
Type
Frameshift
Frameshift
Frameshift
Frameshift
Frameshift
Nonsense
Frameshift
Nonsense
Provean
NA
NA
NA
NA
NA
NA
NA
NA
Frameshift
Single AA Change
Single AA Change
NA
Neutral
Neutral
NA
Damaging
Damaging
Single AA Change Deleterious Damaging
Single AA Change Neutral Damaging
Single AA Change Deleterious Tolerated
Frameshift NA NA
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Single AA Change
Frameshift
Frameshift
Neutral
NA
NA
Damaging
NA
NA
SIFT
NA
NA
NA
NA
NA
NA
NA
NA
Frameshift
Frameshift
Frameshift
NA
NA
NA
NA
NA
NA
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Frameshift NA NA
Single AA Change
Frameshift
Neutral
NA
Tolerated
NA
Single AA Change Deleterious Damaging
Frameshift NA NA
Single AA Change Deleterious Damaging
Nonsense NA NA
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Single AA Change Neutral Damaging
Frameshift
Nonsense
Frameshift
NA
NA
NA
NA
NA
NA
Single AA Change
Frameshift
Single AA Change
Neutral
NA
Neutral
Damaging
NA
Damaging
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Frameshift NA NA
Frameshift
Frameshift
NA
NA
NA
NA
Single AA Change Deleterious Tolerated
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Nonsense NA NA
Nonsense
Frameshift
NA
NA
NA
NA
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Frameshift NA NA
Frameshift
Nonsense
NA
NA
NA
NA
Frameshift
Frameshift
Nonsense
NA
NA
NA
NA
NA
NA
Nonsense NA NA
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Single AA Change Deleterious Tolerated
Insertion Deleterious NA
Single AA Change Deleterious Damaging
Single AA Change Deleterious Damaging
Frameshift NA NA
Frameshift NA NA
Frameshift
Single AA Change
Nonsense
Frameshift
NA
Neutral
NA
NA
NA
Damaging
NA
NA