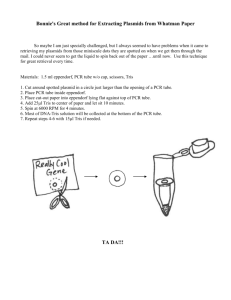
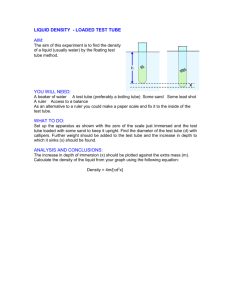
pb150L/stomates
advertisement

Plant Biology 150L Fall 2003 LIGHT-INDUCED PROTEIN EXPRESSION Objectives of this laboratory exercise I. To observe light-induced "greening" of maize seedlings II. To demonstrate the biochemical basis for the greening process Why are we doing this exercise? Light is an important environmental factor that controls plant growth and development. A principal reason is that light makes photosynthesis possible. Besides photosynthesis, light is also involved in other plant processes, mostly affecting the appearance of a plant-- i.e.,, its structural development or morphogenesis (origin of form). The control of morphogenesis by light is called photomorphogenesis. In this lab we will observe how light changes the appearance of darkgrown maize seedlings (Zea maize) and demonstrate the molecular basis of that change. Relevant background information When a seed is sown in darkness (in the soil), it germinates and grows using mostly stored nutrients. Once the seedling penetrates the upper layer of soil and is exposed to light, it begins to alter its form. Perhaps the most striking alteration is the color of the cotyledons (or the leaves) and the stem, from pale yellow to green. This color change is referred to as the "greening" process which prepares the seedling to initiate photosynthesis. Plants can absorb light because they have various photoreceptors, including phytochrome (red far red light), cryptochrome (blue - long wave ultraviolet), UV-B photoreceptor ( UV 280-320 nm), and protochlorophyllide a, which is a precursor of chlorophyll a (red and blue light). The photoreceptor most familiar to us is phytochrome. When phytochrome is exposed to red light, it becomes activated, inducing a series of biochemical changes in the plant cell. One such phytochrome-induced change is the formation of chloroplasts from preplastids. The preplastids already have trace amounts of chlorophyll and proteins required for photosynthesis. Many of the proteins are present at very low levels in dark-grow plants, but upon illumination their expression is dramatically increased. This induction mostly occurs at transcriptional level-i.e., the level of mRNA synthesis. One of the best studied cases in light-induced gene expression is the accumulation of mRNA and protein for ribulose-bisphosphate carboxylase and oxygenase (Rubisco). Studies have shown that the genes coding for the small subunit protein (15 kD) are located in the nucleus while those for the large subunit (50 kD) are in the plastid genome. The small subunit protein is synthesized in the cytoplasm and transported into the plastid. The large subunit is synthesized in the plastid and united with the small subunit, after the latter arrives, to form the holoenzyme. Each functional Rubisco enzyme consists of eight small subunits and eight large subunits. As the enzyme catalyzes the first step in CO2 fixation, Rubisco accumulation rapidly increases during the greening process. Laboratory Procedure 1 Cut 5-10 g (roughly) of etiolated leaves in the dark and store them in liquid N2. 2 The instructor will show you different plants. Compare the plants: dark-grown, greening and green. Examine how significantly light can change the appearance of the plants. Part 1. Light-induced protein synthesis. 3 The instructor will show students how to grind plant tissues in liquid nitrogen. 4 Students will be divided into groups to grind plants and extract proteins from different categories of leaves (etiolated, greening and green). The etiolated leaves are already picked by the instructor before bringing the pots to the lab to avoid illumination. One or two groups can do the etiolated leaves; other groups will pick leaves (5-10 seedlings) from green and “greening” plants and drop them in the mortar containing liquid nitrogen. 5 Grind the tissue into fine powder with a mortar and pestle. 6 Pour the tissue powder into a beaker with 5 ml extraction buffer and mix briefly. 7 Pour into a 15 ml tub, cap the tube and resuspend by inverting several times. 8 Leave the tube on ice for 5 minutes. 9 Take 1 ml of the supernatant and put into an eppendorf tube. Centrifuge for 5 minutes at 14000 rpm at 4°C. This is the crude protein extract stock for further analysis by all groups. 10 Label five eppendorf tubes as “blank,” “control,” “green,” “etiolated,” “greening” (or any other labeling system that will work for you). 11 To each eppendorf tube, add the contents according to the protocol below. Distilled water (DW) is the blank. Bovine Serum Albumin (BSA) is the control. Blank Mix well again by inverting tube. Control Assay Dye. Mix well again by inverting tube. Etiolated Protein Assay Dye. Mix well again by inverting tube. Greening Protein Assay Dye. Mix well again by inverting tube. Green Protein Assay Dye. Mix well again by inverting tube. 12 Measure the absorption at 595 nm in a spectrophotometer. 13 Calculate the protein concentration according to the concentration of the standard (BSA). 14 Compare the results obtained by different groups and take the average if they are close enough (the instructor will make that call). Tube Blank Control Etiolated Greening Green Contents 780 Absorption Protein Concentration Part 2. Protein SDS-PolyAcrylmide Gel Electrophoresis (SDS-PAGE) 1 The gels and running buffer will be made by the instructor during lab. 2 eppendorf tubes, and add an equivalent amount of protein loading buffer to each tube. 3 Heat the samples at 80-90°C for three minutes before loading the MW marker and samples to the gel. 4 Run the gel for 45 minutes at 200 V (vary according to the size of the gel and the position of the dye front). 5 Take off the gel and stain it with Comassie blue for 5-10 minutes. 6 Destain to observe the protein bands around 15 and 50 kD. 7 Compare the abundance of the proteins in different samples Discuss the results according to the following questions: Why are plants changing their color upon illumination? What is the molecular basis for light-induced color change? What other experiments can we do to study the mechanism involved in the light-regulated expression of Rubisco? (e.g., To determine whether the regulation occurs at transcriptional or post-transcriptional level; how the light signal is transmitted to the nucleus; etc.)