lab3_EG
advertisement

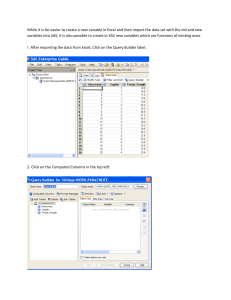
HRP 262, SAS LAB THREE, April 25, 2012 Topics: Cox Regression Lab Objectives After today’s lab you should be able to: 1. Fit models using PROC PHREG. Understand PROC PHREG output. 2. Understand how to implement and interpret different methods for dealing with ties (exact, efron, breslow, discrete). 3. Understand output from the “baseline” statement. 4. Output estimated survivor functions and plot cumulative hazards. 5. Output and plot predicted survivor functions at user-specified levels of the covariates. 6. Understand the role of the strata statement in PROC PHREG. 7. Evaluate PH assumption graphically and by including interactions with time in the model. 8. Use the “where” subsetting statement in all PROC’s. 9. Add time-dependent variables to the model. Understand SAS syntax for time-dependent variables. Be able to correctly specify the time-dependent variables you intend! SAS PROCs PROC LIFETEST PROC LIFEREG PROC PHREG SAS EG equivalent AnalyzeSurvival AnalysisLife Tables None AnalyzeSurvival AnalysisProportional Hazards regression 1 HRP 262, SAS LAB THREE, April 25, 2012 LAB EXERCISE STEPS: Follow along with the computer in front… 1. Go to the class website: www.stanford.edu/~kcobb/courses/hrp262 Lab 2 data SaveSave on your desktop as hmohiv (SAS format) Lab 3 data SaveSave on your desktop as uis (SAS format) 2. Open SAS EG: From the desktop double-click “Applications” double-click SAS Enterprise Guide 4.2 icon 3. Click on “New Project” 4. You DO NOT need to import the dataset into SAS, since the dataset is already in SAS format (.sas7bdat). You DO need to name a library that points to the desktop, where the dataset is located. Assign the library name hrp262 to your desktop folder: ToolsAssign Project Library Name the library HRP262 and then click Next. 2 HRP 262, SAS LAB THREE, April 25, 2012 Browse to find your Desktop. Then Click Next. Click Next through the next screen. Click Finish. 3 HRP 262, SAS LAB THREE, April 25, 2012 5. Confirm that you have created an hrp262 library that contains the SAS datasets uis and hmohiv. 6. We will first use the HMO-HIV dataset to examine ties in a Proportional Hazards model. Recall from last time that the HMOHIV dataset has many ties for time. First we will use the default method (BRESLOW) for dealing with ties. Select AnalyzeSurvival AnalysisProportional Hazards regression Drag time as your Survival time variable and Censor as your censoring variable. Make sure to set the censoring value to 0. 4 HRP 262, SAS LAB THREE, April 25, 2012 Add age and drug as your explanatory variables: Under Methods make sure that you have checked Compute confidence limits for hazard ratio. You will also see the various methods for dealing with failure time ties (Breslow’s approximate likelihood should be chosen by default). 5 HRP 262, SAS LAB THREE, April 25, 2012 Then Click Run. Review of output: 6 HRP 262, SAS LAB THREE, April 25, 2012 Method used for dealing with ties; Breslow is default -2 Log Likelihood for comparing with other models (likelihood ratio test) Test of the global null hypothesis that all coefficients are equal to 0. Wald Tests: tests of whether or not individual betas are equal to 0. 9.6% increase in hazard for every 1-year increase in age, controlling for the effect of IV drug use; 156.3% increase in hazard for using IV drugs, controlling for age. 7. FYI, the corresponding SAS code for proportional hazards regression would be: Syntax for PHREG 7 HRP 262, SAS LAB THREE, April 25, 2012 /**Do not specify ties**/ proc phreg data=hrp262.hmohiv; model time*censor(0)=age drug / risklimits; title 'Cox model for hmohiv data-- ties'; run; risklimits option asks for confidence limits for the estimated hazard ratio. 8. Use the Modify Code button to rerun the Cox model with the other possible specifications for ties (Efron,Exact, Discrete). Then compare. BRESLOW (from above) Analysis of Maximum Likelihood Estimates 95% Hazard Ratio Parameter Standard ChiHazard Confidence Parameter DF Estimate Error Square Pr > ChiSq Ratio Limits Label Age 1 0.09151 0.01849 24.5009 <.0001 1.096 1.057 1.136 Age Drug 1 0.94108 0.25550 13.5662 0.0002 2.563 1.553 4.229 Drug EFRON Analysis of Maximum Likelihood Estimates 95% Hazard Ratio Parameter Standard ChiHazard Confidence Parameter DF Estimate Error Square Pr > ChiSq Ratio Limits Label 8 HRP 262, SAS LAB THREE, April 25, 2012 Analysis of Maximum Likelihood Estimates 95% Hazard Ratio Parameter Standard ChiHazard Confidence Parameter DF Estimate Error Square Pr > ChiSq Ratio Limits Label Age 1 0.09714 0.01864 27.1597 <.0001 1.102 1.062 1.143 Age Drug 1 1.01670 0.25622 15.7459 <.0001 2.764 1.673 4.567 Drug EXACT Analysis of Maximum Likelihood Estimates 95% Hazard Ratio Parameter Standard ChiHazard Confidence Parameter DF Estimate Error Square Pr > ChiSq Ratio Limits Label Age 1 0.09768 0.01874 27.1731 <.0001 1.103 1.063 1.144 Age Drug 1 1.02263 0.25716 15.8132 <.0001 2.781 1.680 4.603 Drug DISCRETE Analysis of Maximum Likelihood Estimates 95% Hazard Ratio Parameter Standard ChiHazard Confidence Parameter DF Estimate Error Square Pr > ChiSq Ratio Limits Label Age 1 0.10315 0.02006 26.4449 <.0001 1.109 1.066 1.153 Age Drug 1 1.07004 0.27438 15.2084 <.0001 2.916 1.703 4.992 Drug Note: only small differences between Breslow, exact, and Efron; Breslow and Efron approximations attenuate slightly toward the null. Discrete gives odds ratios; hence the small increase in size. 9. Corresponding SAS code for different specifications in handling ties. /**Efron**/ proc phreg data=hrp262.hmohiv; model time*censor(0)=age drug / ties=efron risklimits; title 'Cox model for hmohiv data—ties=efron'; run; /**Exact**/ proc phreg data=hrp262.hmohiv; model time*censor(0)=age drug / ties=exact risklimits; title 'Cox model for hmohiv data—ties=exact'; run; /**Discrete**/ proc phreg data=hrp262.hmohiv; model time*censor(0)=age drug / ties=discrete risklimits; title 'Cox model for hmohiv data—ties=discrete'; run; 9 HRP 262, SAS LAB THREE, April 25, 2012 10. Estimate the survival function and plot the cumulative hazard [= -log S(t)]. SAS estimates the baseline hazard and survival functions using a nonparametric maximum likelihood method. Using the estimated coefficients for the covariates (the betas from above), SAS can estimate a survival function for an individual with specific values of the covariates (SAS default plugs in mean values for the cohort). In EG this is as simple as modifying the procedure to estimate the survival function. Click on Modify Task: S (t ) e 0t h ( u ) du log S (t ) 0t h(u )du Go to Results and check the box next to Baseline survivor function estimates. SAS EG will automatically output the estimated survival function (and log survival and log (-log survival) to a temporary dataset. Go to Plots and check Cumulative hazard function plot. 10 HRP 262, SAS LAB THREE, April 25, 2012 Click Run. Under Output Data you should see the following dataset of estimated survival 28 unique event times Mean value of age for the dataset Mean value of drug for the dataset logS(t=2) S (t ) e log(-logS(t=15)) 0t h ( u ) du log S (t ) 0t h(u )du 11 HRP 262, SAS LAB THREE, April 25, 2012 Function curves upward, indicating an increasing hazard with time. FYI, in SAS code we would use a baseline statement to output this estimated survival function (and log survival and log(-log survival)) as follows. : title ' '; proc phreg data=hrp262.hmohiv; model time*censor(0)=age drug / ties=discrete risklimits; baseline out=outdata survival=S logsurv=ls loglogs=lls; run; proc print data=outdata; run; To graph the cumulative hazard using code: data outdata2; set outdata; ls=-ls; run; goptions reset=all; axis1 label=(angle=90); proc gplot data=outdata2; label time='Time(Months)'; label ls='Cumulative Hazard'; plot ls*time / vaxis=axis1; symbol1 value=none i=join; run; 12 HRP 262, SAS LAB THREE, April 25, 2012 11. We can also use these plots to assess the validity of the PH assumption. As discussed in lecture Monday, PH assumption implies that log-log survival curves should be parallel. First we need to rerun the PH analysis to stratify the analysis by the variable Drug. Click on Modify Task: Move Drug from Explanatory variables to Strata variables. Click Run. This should generate a new output dataset containing log(S) and log(-log(S)). Examine the output data -- what is stratification doing? 13 HRP 262, SAS LAB THREE, April 25, 2012 Under your PH analysis, go to the Output Data tab and click on GraphLine Plot. Under Line Plot, choose Multiple line plots by group column Under Data, plot time on the Horizontal by loglogsurvival on the Vertical. Group the plots by Drug. 14 HRP 262, SAS LAB THREE, April 25, 2012 Click Run. FYI, the SAS code equivalent is as follows: Note that DRUG has been removed from the model statement; stratifying by DRUG allows SAS to assume different baseline hazards for each drug group (which can later be compared to test PH assumption)… proc phreg data=hrp262.hmohiv; model time*censor(0)= age/ ties=discrete risklimits; strata drug; 15 HRP 262, SAS LAB THREE, April 25, 2012 baseline out=outdata survival=S logsurv=ls loglogs=lls; run; **Use SolutionsAnalysisInteractive Data AnalysisWork.Outdata to examine the contents of outdata. What is stratification doing? proc gplot data=outdata; title 'Evaluate proportional hazards assumption for variable: drug'; plot lls*time=drug /vaxis=axis1; symbol1 i=join c=black line=1; symbol2 i=join c=black line=2; run; log Si(t ) log S j (t ) HR log Si (t ) HR log S j (t ) becauselog no events log( log Si (t )) log(Short HR S j (t )) in this group beyond t=20 log( log Si (t )) log HR log( log S j (t )) X (t ) K X (t ) Recall: in SAS LAB 2 we graphed log(-log)S(t) for these data against log(time) using proc lifetest, which involved fewer steps!! 16 HRP 262, SAS LAB THREE, April 25, 2012 10. Since it’s hard to tell if the hazards are proportional, we might wonder if PH assumption is violated and if there is an interaction between drug and time. We can test this (and fix problem) by adding such an interaction term to the model. **We have to use code to do this, because “time” is a time-changing variable; thus we cannot simply create a time*drug variable and add that to the model. The drugtime variable that we are creating below is a time-dependent variable, where “time” is updated at every event time (SAS does this work for us). Find the previous Cox regression run that included age and drug as predictors. Directly modify the code as follows: PROC PHREG DATA=WORK.TMP0TempTableInput PLOTS=SURVIVAL ; MODEL time * Censor (0) = Drug Age drugtime / TIES=BRESLOW RISKLIMITS ALPHA=0.05 SELECTION=NONE ; drugtime=drug*time; RUN;TITLE; SAS automatically creates timedependent variables for you in the phreg procedure (without a separate datastep). This temporary variable is not saved. Analysis of Maximum Likelihood Estimates 95% Hazard Ratio Parameter Standard ChiHazard Confidence Parameter DF Estimate Error Square Pr > ChiSq Ratio Limits Label Drug 1 1.12079 0.34996 10.2570 0.0014 3.067 1.545 6.090 Drug Age 1 0.08925 0.01877 22.6193 <.0001 1.093 1.054 1.134 Age drugtime 1 -0.02818 0.03884 0.5264 0.4681 0.972 0.901 1.049 p-value is not significant, so there is little evidence of an interaction exists. 11. Obtain predictions about survival times for particular sets of covariate values that need not appear in the data set being analyzed. You have to create a new dataset that contains the covariate values of interest. We’ll use code to enter the new dataset (though this could also be done in point and click). NewNew Program 17 HRP 262, SAS LAB THREE, April 25, 2012 data MyCovs; input age drug; datalines; 55 1 22 0 ; run; Then run a Cox regression model on the original hmohiv dataset, with age and drug as predictors: Drag time as your Survival time variable and Censor as your censoring variable. Make sure to set the censoring value to 0. Add age and drug as your explanatory variables: 18 HRP 262, SAS LAB THREE, April 25, 2012 Under Resultsask for Baseline survivor function estimates; Then click Browse to rename these data… Find the Work libraryName the dataset Outdata 19 HRP 262, SAS LAB THREE, April 25, 2012 Then click Save and Run. 20 HRP 262, SAS LAB THREE, April 25, 2012 Examine the data under Output Data: Defines the survivor curve at the mean age and mean IV drug use. Make one modification to the code to apply your model to the MyCovs dataset (e.g., to get survivor curves for a 55-year old drug user and a 22-year old non-drug user). BASELINE OUT=WORK.OUTDATA(LABEL="Baseline Survivor Function Estimates for WORK.QUERY_FOR_HMOHIV_0000") SURVIVAL=_SURVIV_ UPPER=_SDFUCL_ LOWER=_SDFLCL_ LOGLOGS=_LOGLOGS_ LOGSURV=_LOGSURV_ STDERR=_STDERR_ STDXBETA=_STDXBETA_ XBETA=_XBETA_ covariates=mycovs ; 21 HRP 262, SAS LAB THREE, April 25, 2012 RUN; Note the output data now contains two survivor curves, one for a 55 year old drug user and 1 for a 22 year old non-drug user: 12. Graph these survival curves (we’ll use code here so that we can superimpose the confidence limits on the same graph): goptions reset=all; axis1 label=(angle=90); proc gplot data=outdata; title 'Survivor f3unction at age 55 with drug'; plot _surviv_*time _sdflcl_*time _sdfucl_*time/overlay vaxis=axis1; symbol1 i=join c=black line=1; Note use of the very convenient symbol2 i=join c=black line=2; where statement (works in most symbol3 i=join c=black line=2; SAS procs) for subsetting. where age=55; run; proc gplot data=outdata; title 'Survivor function at age 22 without drug'; plot _surviv_*time _sdflcl_*time _sdfucl_*time /overlay vaxis=axis1; symbol1 i=join c=black line=1; symbol2 i=join c=black line=2; symbol3 i=join c=black line=2; where age=22; run; 22 HRP 262, SAS LAB THREE, April 25, 2012 It’s a quick drop-off for this group. Note that the confidence intervals go haywire around PredictedSurvival=0. 23 HRP 262, SAS LAB THREE, April 25, 2012 Now, let’s switch datasets to UIS. The data dictionary is below for your reference: These data were collected from a randomized trial of two in-patient treatment courses for drug addiction (heroin, cocaine, and unspecified other drugs): a shorter treatment plan and a longer treatment plan. Time to censoring or return to drug use was recorded, as was length of time in the treatment plan. Variables are below: Variable ID AGE BECKTOTA Description Identification code Age at enrollment Beck Depression Score at Admission Heroin/Cocaine use during 3 months prior to admission Codes/Values 1-628 Years 0-54 IVHX IV drug use history at admission NDRUGTX Number of prior drug treatments Subject’s race 1=never 2=previous 3=recent 0-40 HERCOC RACE TREAT SITE LOT TIME CENSOR Treatment randomization Assignment Treatment site Length of treatment (exit date-admission date) Time to return of drug use (Measured from admission) Returned to drug use 1=Heroin& cocaine 2=Heroin only 3=Cocaine Only 4=neither 0=White 1=Other 0=short 1=long 0=A 1=B Days Days 1=Returned to drug use 0=Otherwise 24 HRP 262, SAS LAB THREE, April 25, 2012 14. Examine data using a KM plot stratified by “TREAT” which specifies the randomization group (short or long). In EG, open the UIS dataset. Click on AnalyzeSurvival Analysis Life Tables. Drag time to Survival time, censor as the Censoring variable, and treat as the Strata variable. Make sure to specify 0 as the Censoring value. Click Run. 25 HRP 262, SAS LAB THREE, April 25, 2012 Scroll to the bottom of the results window to examine the KM plot. The analogous SAS code is: goptions reset=all; Note use of proc format to assign proc format; format to the variable treat. value treat 1 = "long" 0 = "short"; run; proc lifetest data=hrp262.uis plots=(s) graphics censoredsymbol=none; label time='time(days)'; time time*censor(0); strata treat; title 'UIS KM plot'; symbol1 c=black v=none line=1 i=join; symbol2 c=black v=none line=2 i=join; *gives plots that will be suitable for black and white copying; format treat treat.; run; 15. Now run a PH analysis with treat and age as predictors, first as a crude analysis and then stratified by site. Scroll through and review output as a class. 26 HRP 262, SAS LAB THREE, April 25, 2012 Under the UIS dataset, click on AnalyzeSurvival AnalysisProportional Hazards. For the non-stratified analysis, specify time under Survival time, censor as the Censoring variable, age and treat as the Explanatory variables. Click Run. WITHOUT STRATIFICATION BY SITE: Analysis of Maximum Likelihood Estimates Interpretation: there is a 20% decre relapse in those treated in the long 27 those in the short p compared with HRP 262, SAS LAB THREE, April 25, 2012 Parameter Standard Hazard Parameter DF Estimate Error Chi-Square Pr > ChiSq Ratio age 1 -0.01327 0.00721 3.3847 0.0658 0.987 treat 1 -0.22298 0.08933 6.2307 0.0126 0.800 Now repeat the analysis with site as the Stratification variable: Click Run. WITH STRATIFICATION BY SITE: Analysis of Maximum Likelihood Estimates Parameter Standard Hazard Parameter DF Estimate Error Chi-Square Pr > ChiSq Ratio age 1 -0.01429 0.00729 3.8434 0.0499 0.986 treat 1 -0.23507 0.08961 6.8820 0.0087 0.791 The fact that there’s little difference between the hazard ratios with or without stratification indicates that there’s little confounding by site, BUT it might be important to leave the stratification in anyway, to avert potential criticism by reviewers of your analysis. There seems to be a protective effect for being randomized to the longer treatment group. Stratification allows there to be totally different (and even non-parallel) baseline hazard functions in each stratum. Rather than trying to estimate hazard ratios between the strata, you instead generate a hazard ratio that has been “averaged” over the different strata (like a mantel-haenszel odds ratio). Risk sets for the partial likelihood are allowed to be stratum-specific. This is useful if you want to control for a possible confounding variable that (1) may violate the Proportional Hazards assumption and/or (2) is a nuisance variables—that is you don’t care about getting hazard ratios for it. For example, in this study, there were two hospitals in which treatment was administered (site 1 and site 2). There’s good reason to believe that baseline hazards might be totally different depending on the site (because of different local populations, and different ways of administering the treatment). For this study, we don’t care about the effect of site; we just want to know if the long treatment is better than the short treatment. So, we “average” the HR for the long treatment compared with the short treatment over the two different sites. 28 HRP 262, SAS LAB THREE, April 25, 2012 The corresponding SAS code is: Without stratification: proc phreg data=hrp262.uis; model time*censor(0)=age treat/risklimits; run; With stratification: proc phreg data=hrp262.uis; model time*censor(0)=age treat/risklimits; strata site; run; 16. Run PROC PHREG with treatment as a time-dependent variable and age (much easier with code). proc phreg data=hrp262.uis; model time*censor(0)=off_trt age treat/risklimits; if lot>=time then off_trt=0; else if lot<time then off_trt=1; run; This is adding a time-dependent covariate: off_trt, which is 1 if they have left the treatment program and 0 if they are still in the treatment program. This is evaluating whether just being in the treatment facility and program prevents a relapse. At each event time T, for each individual still in the risk set, SAS compares the length of treatment to T. For example, the first event occurred at 2 months. SAS goes through and compares the length of treatment for each individual with 2 months. If the person was in treatment longer than 2 months, then they are still on treatment at the first event time and off_trt=0. If the person was in treatment shorter than 2 months, then at 2 months they are off treatment and off_trt=1. Thus, the values of off_trt change at each event time and in each likelihood term of the likelihood equation. RESULTS: Analysis of Maximum Likelihood Estimates Variable DF Parameter Estimate Standard Error Chi-Square Pr > ChiSq Hazard Ratio off_trt age treat 1 1 1 2.56681 -0.00719 0.01082 0.15173 0.00728 0.08988 286.1826 0.9777 0.0145 <.0001 0.3228 0.9042 13.024 0.993 1.011 95% Hazard Ratio Confidence Limits 9.674 0.979 0.848 17.535 1.007 1.206 There appears to be no difference between short vs. long treatment, beyond the fact that currently being in treatment prevents relapse and the longer program keeps them in longer. 29