Supplementary Data
advertisement

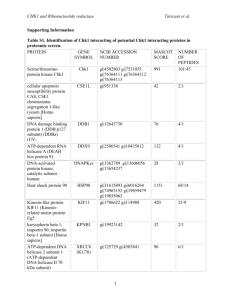
Online Appendix for the following JACC article TITLE: Provocation of an Autoimmune Response to Cardiac Voltage-Gated Sodium Channel NaV1.5 and Induces Cardiac Conduction Defects in Rats AUTHORS: Sevil Korkmaz, PHD, Edgar Zitron, MD, Anna Bangert, Claudia Seyler, PHD, Shiliang Li, MD, Peter Hegedüs, MD, Daniel Scherer, MD, Jin Li, MD, Thomas Fink, Patrick A. Schweizer, MD, Evangelos Giannitsis, MD, Matthias Karck, MD, Gábor Szabó, MD, PHD, Hugo A. Katus, MD, Ziya Kaya, MD APPENDIX Supplementary Data Online Methods Enzyme-linked immunosorbent assay (ELISA) for autoantibodies detection Serum samples were obtained after centrifugation of blood collected from rats on days 0, 7, 14, 21 and 28. Antibody titers were determined using ELISA technique, as described previously [1]. Briefly, 96-well plates were coated with 5µg/mL NaV1.5-peptide (100µL/well) dissolved in bicarbonate buffer (pH 9.6) and incubated overnight at 4°C. After 3 washing steps (using 1xPBS/Tween20), plates were then blocked with 1%BSA/1xPBS overnight at 4°C. Serum samples were serially diluted to 1:200, 1:800, 1:3200, 1:12800, and 1:204800 and pipetted onto the plates for 3 hours at room temperature. Normal rat serum was used as control. Horseradish peroxidase (HRP)-coupled goat anti-rat secondary antibody (Sigma, St. Louis, MO, USA) diluted to 1:5000 was used for detection. The peroxidase activity was detected with 3,3',5,5'-tetramethylbenzidine (TMB). Optical densities were determined at 450 nm. Antibody end-point titer for each individual rat was calculated as the greatest positive dilution of antibody that yielded a 1 positive signal. Electrocardiography 12-lead ECG was recorded for 1 hour on days 21 and 28 after first immunization. Rats were anesthetized intraperitoneally with a mixture of ketamine (100mg/kg) and xylazine (3mg/kg) and kept in a supine position on heating pads maintaining their core temperature (measured via a rectal probe) at 37°C. Standard 12-lead ECG was recorded using needle electrodes placed subcutaneously. All leads were connected to a standard direct-writing recorder (Mortara Instrument, WI, USA). Paper speed was set at 50 mm/s and the sensitivity was 10 mm/mV. The ECG analysis was evaluated in lead II including the following measurements: PR-interval, QRS complex duration, ST-segment elevation, and QT-interval. QT-interval, measured from the onset of the QRS complex to the end of the T wave, was corrected using normalized Bazett’s formula adjusted for rats (nQTc=QT/(RR/f)1/2) [2]. ECG-recordings were analyzed by an investigator blinded to the experimental groups. Telemetry-ECG On day 25 after first immunization, rats were anesthetized intraperitoneally with a mixture of ketamine (100mg/kg) and xylazine (3mg/kg). A midline abdominal incision was made and the transmitter devices (ECG-Device; Data Sciences International) were placed under subcutaneous tissue of the abdomen, the leads were tunneled under the skin to the recording sites and attached to the underlying tissue to prevent migration. The abdominal skin was closed and rats were kept on a heating pad (37-38°C) until that they were fully recovered from anesthesia. Analgesia was achieved by the administration of buprenorphine (0.05mg/kg, s.c.; every 12 hours for 2 days). After a period of 3 days, the transmitters in the animals were activated by use of a magnet switch and signals were collected by a receiver placed under the cage for a period of 24 hours. In vivo hemodynamic measurements 2 On day 28 after first immunization, rats were anesthetized with sodium pentobarbital (60 mg/kg i.p.). Animals were placed on heating pads and the core temperature monitored through rectal probe was maintained at 37°C as described [3]. After tracheotomy, rats were intubated and ventilated. A polyethylene catheter was inserted into the left external jugular vein for fluid administration. A 2F microtip pressure-volume catheter (SPR-838, Millar Instruments, Houston, TX, USA) was inserted into the right carotid artery and advanced into the ascending aorta. After stabilization for 5 minutes, arterial blood pressure was recorded and the catheter was advanced into the left-ventricle under pressure control. With the use of a special pressure-volume analysis program (PVAN, Millar Instruments, Houston, TX, USA), heart rate, systolic and diastolic blood pressures, mean arterial pressure, left-ventricular (LV) endsystolic pressure, LV end-diastolic pressure, end-systolic volume, end-diastolic volume, stroke volume, maximal slope of the systolic pressure increment (dP/dtmax), maximal slope of the diastolic pressure decrement (dP/dtmin), ejection fraction, and cardiac output were calculated. Ventricular relaxation was assessed by the time constant of LV pressure decay (Tau) and has been calculated by the Glantz method (Tau-g; regression of dP/dt versus pressure). LV pressure-volume relations were assessed by transiently compressing the inferior vena cava. The slope of the dP/dt max-end-diastolic volume, and preload recruitable stroke work (PRSW) were calculated as load-independent indexes of LV contractility. The slope of the LV end-diastolic pressure-volume relationship (EDPVR) was calculated as a reliable index of LV stiffness. At the end of each experiment, 0.1 mL hypertonic saline was injected using the central venous line, and from the shift of pressure-volume relations parallel conductance volume was calculated by pressurevolume analysis software (PVAN, Millar Instruments, Houston, TX, USA) and used for correction of absolute LV volume. The volume calibration of the conductance system was performed as described previously [4]. Nine cylindrical holes in a block with 10 mm depth and a diameter ranging from 2 to 11 mm were filled with fresh heparinized rat blood. The linear volume-conductance regression of the absolute volume in each cylinder in comparison to the raw signal acquired by the conductance catheter was used as the volume calibration formula. 3 Histopathology Dissected sinoatrial and atrioventricular nodes and longitudinal sections were fixed in buffered paraformaldehyde solution (4%), and embedded in paraffin. We performed serial paraffin heart sections with a thickness of 3-µm. Five sections per heart were inspected under light microscopy and rated according to following scoring system: Grade 1, cardiac infiltration in up to 5% of the cardiac section; Grade 2, infiltration in 6-10%; Grade 3, infiltration in 11-30%; Grade 4, infiltration in 31-50%; and Grade 5, infiltration >50% of the cardiac section. Histopathologic evaluation was conducted by an examiner blinded to the treatment groups. Quantitative polymerase chain reaction (PCR) Total RNA was extracted from heart with the RNAeasy Minikit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Quantitative PCR for NaV1.5, tissue inhibitor of metalloproteinase (TIMP)-1, matrix metalloproteinases (MMP)-2, MMP-9, and MMP-14 were carried out with 0.5 µL of cDNA, assembled from 1 µg RNA in a 20 µL reaction by using the iScript Kit (Bio-Rad Laboratories, Munich, Germany). After an initial denaturation at 95°C for 30 seconds, 40 cycles consisting of denaturation at 95°C for 5 seconds and annealing at 60°C for 10 seconds followed. Primer sequences used for quantification of transcript levels were as follows: NaV1.5: forward: 5’-tgccaccaatgccttgta-3’; reverse: 5’catgatgagcatgctaaagagc-3’; TIMP-1: forward: 5’-cagcaaaaggccttcgtaaa-3’; reverse: 5’- tggctgaacagggaaacact-3’; MMP-2: forward: 5’-caccaccgaggattatgacc-3’; reverse: 5’- cacccacagtggacatagca-3’; MMP-9: forward: 5’-cctctgcatgaagacgacataa-3’; reverse: 5’- ggtcaggtttagagccacga-3’; MMP-14: forward: 5’-ggactgtccaaaatgaggatct-3’; reverse: 5’- 5’-ggagaagaaaccctcacaagc-3’; reverse: 5’- gaggggtcgttggaatgtt-3’; and L-32: forward: gccacaaaggacgtgtttc-3’. Quantification was done using the Quanti Fast Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The ribosomal protein L32 served as reference gene. 4 Western Blot Myocardial proteins were extracted by using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen, Hilden, Germany). The protein pellet was dissolved in 8M urea and the concentration was determined by a commercial kit according to the manufacturer’s protocol (Bradford Bio-Rad Protein Assay, Bio-Rad, München, Germany). Total protein homogenates (10μg) were denatured, separated on sodium dodecyl sulfate polyacrylamide gel electrophoresis gradient gels (Invitrogen, Darmstadt, Germany) and transferred to polyvinylidene fluoride membrane (Invitrogen, Darmstadt, Germany). The membranes were blocked with 5% milk in Tris-Buffered Saline Tween20 before incubation overnight at 4°C with pooled serum from NaV1.5-immunized rats (1:1000) or primary antibodies specific to NaV1.5 (1:750, Abcam, Cambridge, UK). After washing blots to remove excessive primary antibody binding, blots were incubated for 1h with horseradish peroxidase conjugated secondary antibodies (1:10000, Sigma-Aldrich, Steinheim, Germany). The immunoreactive protein bands were developed using an Enhanced Chemiluminescence system (GE Healthcare Europa GmbH, Freiburg, Germany). The intensity of immunoblot bands was detected with Hyperfilm™ ECL (GE Healthcare Europa GmbH, Freiburg, Germany). Ventricular cardiomyocyte isolation and treatment Adult ventricular cardiomyocytes were isolated with methods described previously [5]. Briefly, rats were anesthetized with sodium pentobarbital (60 mg/kg i.p.), heparinized (500 UI/kg), the hearts were rapidly removed, washed in cold 0.9% NaCl solution, then perfused in a Langendorff perfusion system at 37°C with culture medium containing EDTA and subsequently digested by collagenase type II (240 U/mg; Worthington Biochemical Corporation, NJ, USA) to break down the extracellular matrix. Then, the ventricles were minced and the resulting cellular digest was washed, filtered through a 500 µm mesh (Corning, NY, USA), and resuspended in supplemented Medium 199 (Sigma Aldrich Chemie GmbH, Germany). Cells were allowed to sediment by gravity and calcium concentration was gradually increased in several steps which allow the cells to return to normal cytosolic calcium levels. Freshly isolated 5 cardiomyocytes were attached to laminin coated 25 mm cover slips by 1 hour incubation at 37°C and 5% CO2. After 1 hour, the medium was changed to a medium containing 1:10 diluted sera from control or NaV1.5-immunized rats, and then incubated again for at least 1 hour. Whole-cell patch clamp recordings Measurement of INa and Ito Patch-clamp measurements of rat ventricular cardiomyocytes were performed in a bath with a lowsodium solution (in mmol/L): NaCl 20.0, N-methyl-D-glucamine 130.0, KCl 5.4, HEPES 10.0, CdCl2 0.1, MgCl2 2.0, CaCl2 0.5, BaCl2 2.0, glucose 10.0, pH adjusted to 7.4 with HCl. 10 µmol/L nisoldipine to block calcium currents, 10 µmol/L chromanol 293B to inhibit the slow component IKs of the delayed rectifier potassium current, 10 µmol/L E-4031 to block the rapid component IKr of the delayed rectifier potassium current, and 1 µmol/L glibenclamide to inhibit the ATP-dependent IK(ATP) were added to the bath solution directly before the measurements. Electrodes were filled with a solution containing (in mmol/L): KCl 140, MgCl2 4.0, CaCl2 5.0, EGTA 10.0, HEPES 10.0, and Na2-ATP 5.0, so that the transmembrane Na+ gradient was 10 mmol/L, and adjusted to pH 7.3 with KOH. In order to measure current-voltage relations (range -60 to +20 mV), 20-ms clamp steps were applied in 10-mV increments. For steady-state inactivation (range -140 to -20 mV), 400-ms conditioning clamp steps (10-mV increments) were followed by a test clamp step to -20 mV (duration 20 ms); a step of 2 ms at -100 mV was interposed between conditioning and test clamp steps. Small cells were chosen to minimize voltage clamp errors. The scientists performing and analyzing the electrophysiological measurements were blinded with respect to the identity of the applied sera. Only single rod-shaped myocytes showing clear striations were selected for the experiments. Sodium currents were recorded with the whole cell patch-clamp technique. Currents were measured with an RK400 amplifier (Bio-Logic SAS, Claix, France), stored on hard disk, and analyzed with pCLAMP (Axon Instruments, Foster City, USA) software. The patch pipettes pulled from borosilicate glass (1B120F-4; 6 World Precision Instruments, Berlin, Germany) were generated on a Flaming/Brown micropipette puller P-87 (Sutter Instruments, Novato, USA) and fire polished to give a final resistance of 1.5-2.5 MΩ. Recordings with less than 10% leak current were considered for data analysis. All experiments were performed at room temperature (20-24°C) to improve both stability of myocytes and voltage control when measuring rapidly activating currents. No leak subtraction was performed during the experiments. Measurement of L-type ICa To record ICa,L, a prepulse (25 ms) was applied to -40 mV from a holding potential of -80 mV to inactivate the Na+ channels and the T-type Ca2+ channels, followed by voltage steps (300 ms) from -40 mV to +60 mV in 10 mV increments. Contaminating K+ currents were blocked by replacing K+ with Cs+. The superfusion solution was composed of (in mM): tetraethylammonium (TEA) chloride 136, HEPES 10, CaCl2 2, MgCl2 2 and glucose 10, pH 7.4 (adjusted with TEA-OH). The pipette solution (pH 7.2 with Cs-OH) included (in mM): cesium chloride 140, HEPES 10, Mg–ATP 2 and EGTA 10. The tip resistances of the pipettes were 2-3 MΩ. Patients with idiopathic atrioventricular block The following pre-defined 168 antigens have been used for microarray analysis: Myosin-binding protein C, cardiac-type (Q14896), Myosin-7 (P12883), Troponin T, cardiac muscle (P45379), Troponin I, cardiac muscle (P19429), Tropomyosin alpha-1 chain (P09493), Myosin light chain 3 (P08590), Myosin regulatory light chain 2 ventricular/cardiac muscle isoform (P10916), Serine/threonine-protein kinase TNNI3K (Q59H18), Actin, alpha cardiac muscle 1 (P68032), Titin (Q8WZ42), Troponin C, slow skeletal and cardiac muscles (P63316), Myosin-6 (P13533), Cysteine and glycine-rich protein 3 (P50461), Caveolin-3 (P56539), Telethonin (O15273), Vinculin (P18206), Junctophilin-2 (Q9BR39), Obscurin (Q5VST9), Myozenin-2 (Q9NPC6), Ankyrin repeat domaincontaining protein 1 (Q15327), Desmin (P17661), Prelamin-A/C (P02545), Delta-sarcoglycan (Q92629), Alpha-actinin-2 (P35609), LIM domain-binding protein 3 (O75112), Cardiac phospholamban (P26678), 7 ATP-binding cassette sub-family C member 9 (O60706), Sodium channel protein type 5 subunit alpha (Q14524), Alpha-crystallin B chain (P02511), Presenilin-1 (P49768), Presenilin-2 (P49810), Four and a half LIM domains protein 2 (Q14192), Integrin-linked protein kinase (Q13418), Myopalladin (Q86TC9), Muscarinic acetylcholine receptor M2 (P08172), Dystrophin (P11532), Emerin (P50402), Tafazzin (Q16635), Fukutin (O75072), Desmoplakin (P15924), Junction plakoglobin (P14923), Plakophilin-2 (Q99959), Transforming growth factor beta-3 (P10600), Ryanodine receptor 2 (Q92736), Desmoglein-3 (P32926), Dystrobrevin alpha (Q9Y4J8), Glycogen synthase kinase-3 alpha (P49840), Glycogen synthase kinase-3 beta (P49841), High mobility group protein B1 (P09429), Dual specificity tyrosinephosphorylation-regulated kinase 1A (Q13627), Leptin (P41159), Regulator of G-protein signaling 10 (O43665), Regulator of G-protein signaling 2 (P41220), Caveolin-2 (P51636), Annexin A2 (P07355), Cathepsin B (P07858), Transmembrane protein 43 (Q9BTV4), Lysosome-associated membrane glycoprotein 2 (P13473), Alpha-galactosidase A (P06280), 5'-AMP-activated protein kinase catalytic subunit alpha-2 (P54646), E3 ubiquitin-protein ligase TRIM21 (P19474), BTB/POZ domain-containing protein KCTD14 (Q9BQ13), Protein S100-A1 (P23297), TIR domain-containing adapter molecule 1 (Q8IUC6), Vascular cell adhesion protein 1 (P19320), Intercellular adhesion molecule 1 (P05362), Eselectin (P16581), Plasminogen activator inhibitor 1 (P05121), Nexilin (Q0ZGT2), Beta-1 adrenergic receptor (P08588), Calmodulin (P62158), E3 ubiquitin-protein ligase TRIM63 (Q969Q1), Myozenin-2 (Q9NPC6), 5'-AMP-activated protein kinase subunit gamma-2 (Q9UGJ0), Ankyrin-2 (Q01484), Sodium channel subunit beta-4 (Q8IWT1), Voltage-dependent L-type calcium channel subunit alpha-1C (Q13936), Inward rectifier potassium channel 2 (P63252), Potassium voltage-gated channel subfamily E member 2 (Q9Y6J6), Potassium voltage-gated channel subfamily KQT member 1 (P51787), Voltagedependent anion-selective channel protein 1 (P21796), Alpha-enolase (P06733), Serpin H1 (P50454), Phenylethanolamine N-methyltransferase (P11086), POU domain, class 5 transcription factor 1 (Q01860), Homeobox protein Nkx-2.5 (P52952), Myocyte-specific enhancer factor 2C (Q06413), Transcription factor GATA-4 (P43694), Heat shock protein beta-1 (P04792), Heat shock protein beta-3 (Q12988), 60 kDa heat shock protein, mitochondrial (P10809), Heat shock protein HSP 90-alpha (P07900), Heat shock 8 protein HSP 90-beta (P08238), 5-hydroxytryptamine receptor 4 (Q13639), Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (O14983), Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (P16615), Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (Q93084), Annexin A5 (P08758), Annexin A6 (P08133), Natriuretic peptides A (P01160), Heparin-binding growth factor 2 (P09038), Natriuretic peptides B (P16860), 2',3'-cyclic-nucleotide 3'-phosphodiesterase (P09543), Gap junction alpha-1 protein (P17302), Heart- and neural crest derivatives-expressed protein 2 (P61296), Heart- and neural crest derivatives-expressed protein 1 (O96004), Transcription factor GATA-6 (Q92908), Fatty acid-binding protein, heart (P05413), Insulin-like growth factor I (P05019), Laminin subunit gamma-1 (P11047), Laminin subunit alpha-5 (O15230), Laminin subunit alpha-4 (Q16363), Laminin subunit alpha2 (P24043), Laminin subunit beta-1 (P07942), Laminin subunit beta-2 (P55268), Laminin subunit alpha-1 (P25391), Laminin subunit gamma-3 (Q9Y6N6), Laminin subunit beta-4 (A4D0S4), Laminin subunit beta-3 (Q13751), Laminin subunit alpha-3 (Q16787), Laminin subunit gamma-2 (Q13753), Sodium/potassium-transporting ATPase subunit alpha-2 (P50993), Sodium/potassium-transporting ATPase subunit alpha-3 (P13637), Protein Wnt-7a (O00755), Protein Wnt-10b (O00744), Protein Wnt10a (Q9GZT5), Protein Wnt-8b (Q93098), Protein Wnt-3a (P56704), Protein Wnt-9b (O14905), Protein Wnt-2b (Q93097), Protein Wnt-16 (Q9UBV4), Protein Wnt-11 (O96014), Protein Wnt-9a (O14904), Protein Wnt-5a (P41221), Protein Wnt-4 (P56705), Protein Wnt-5b (Q9H1J7), Protein Wnt-7b (P56706), Protein Wnt-6 (Q9Y6F9), Protein Wnt-8a (Q9H1J5), Protein Wnt-2 (P09544), Proto-oncogene Wnt-3 (P56703), Proto-oncogene Wnt-1 (P04628), Plakophilin-1 (Q13835), Plakophilin-2 (Q99959), Plakophilin-3 (Q9Y446), Plakophilin-4 (Q99569), Glycogen synthase kinase-3 beta (P49841), Plateletderived growth factor subunit A (P04085), Platelet-derived growth factor subunit B (P01127), Plateletderived growth factor C (Q9NRA1), Platelet-derived growth factor D (Q9GZP0), Cadherin-2 (P19022), Cadherin-12 (P55289), Metalloproteinase inhibitor 1 (P01033), Metalloproteinase inhibitor 3 (P35625), Metalloproteinase inhibitor 2 (P16035), Metalloproteinase inhibitor 4 (Q99727), Creatine kinase U-type, mitochondrial (P12532), Myosin regulatory light chain 2 atrial isoform (Q01449), Myosin light chain kinase, smooth muscle (Q15746), Cardiac myosin light chain-1 (9651188), Cardiac myosin light chain 2 9 (1220301), Endothelin-1 (P05305), ADM (P35318), BAG family molecular chaperone regulator 3 (O95817), Eyes absent homolog 4 (O95677), Nebulette (O76041), Calreticulin-3 (Q96L12). The antigen sequences were linked together to form a single polyprotein sequence; the C- and N-terminus of polyprotein were elongated by neutral GS linkers to avoid truncated peptides. The full sequence was then translated into 15mer peptides in duplicates with a peptide-peptide overlap of 10 amino acids. Translation of the linkage sites of adjacent proteins resulted in artificial peptides with sequence contributions of both proteins. The peptide microarray array was framed by Flag (DYKDDDDKGG, 2.167 spots) and HA (YPYDVPDYAG, 2.149 spots) control peptides. After 10 min pre-swelling in standard buffer and 60 min in blocking buffer, the peptide microarrays were initially incubated only with the secondary antibody at a dilution of 1:5000 for 60 min at room temperature to analyze background interactions with the polyprotein-derived 15mer peptides. At a scanning intensity of 7, we observed some medium intensity interactions with arginine- and lysine-rich peptides (e.g. SIQISDKTKRKMRRR, GPRGLQATARKASRR, KKLSKDRMKKYMARR and KKDGDKKKKKKIKEK as most intensive binders) due to non-specific ion-exchange effects of the antibody dye with this strongly basic motifs (see Excel file Peptide_Map_PreStaining.xlsx; raw data not shown). Some higher intensity spots with deviations of >50% were regarded as artifacts. After 2x1 min washing and 30 min swelling in standard buffer, the peptide microarrays were incubated overnight at 4°C with the human serum samples AV Block and Normal (alt) at a dilution of 1:1000 and shaking at 500 rpm. Repeated washing in standard buffer (2x10 sec and 1x1 min) was followed by incubation for 30 min with the secondary F(ab´)2 goat anti-human IgG(H+L) conj. DyLight680 antibody at a dilution of 1:5000 at room temperature. After 2x1 min washing in standard buffer, the microarray was rinsed with Millipore® water and dried in a stream of air. Read-out was done with an Odyssey Imaging System at a resolution of 21 μm and green/red intensities of 7/7 before and after staining of control antibodies. 10 Staining of Flag and HA control peptides that frame the arrays gave rise to high and homogeneous spot intensities. All serum samples showed complex staining patterns indicative for a polyclonal response. Despite the pooled nature of the serum samples, the signal to noise ratio was surprisingly good with a comparably low background. The antibody responses seemed to be differential for all samples with control serum showing the weakest response. A row of double spots with strongly acidic peptides (EKSKKKKEEEEDEED to EDEDEEEDDDDEMHT) was reactive in all samples due to ionic interactions with serum antibodies. Such antibody-peptide interactions were regarded as non-specific and not discussed in detail. Spot Quantification and Data Evaluation Quantification of spot intensities and peptide annotation were done with PepSlide® Analyzer before staining of the control antibodies. A software algorithm breaks down fluorescence intensities of each spot into foreground and background signal. The foreground median intensities of each peptide were averaged over the double spots and the standard deviation calculated. Based on averaged median intensities, an intensity map was generated to correlate the microarray layout shown in the peptide map with intensity data. To facilitate differential analysis of serum responses, the most intensive interactions with control serum were also highlighted by a color code. Data normalization and statistical analysis were omitted at this stage. We additionally calculated OD values for all patient group samples by dividing a given peptide intensity by the corresponding intensity measured in control serum sample. Peptide and intensity maps as well as the mapping summaries were finally correlated with visual inspection of the microarray scans to identify hot spots and predominant peptide-serum interactions that hint at immunogenic sites in the 168 antigens investigated in line of this discovery screen. In line of visual inspection, we focused on a row of two or three peptide double spots that may contain a consensus motif as epitope. The unambiguous identification of consensus motifs formed by neighbored peptides was further supported by MEME Suite (Motif-based sequence analysis tools). For MEME analysis we set cut- 11 off level of the 10fold average foreground intensities of all spots, defined a minimal motif length of 4 amino acids and a maximum output of different 20 motifs. 12 13 Online Table 1. Microarray results of autoantibodies in the sera from the patients with idiopathic atrioventricular block (AVB) identifying different peptide consensus motifs of the NaV1.5. *Amino acid residues corresponding to the third extracellular loop of the NaV1.5, between the fifth and the sixth transmembrane Amino acid residues Ratio AVB/mean Normal Intensity segment of the first domain. (>3.00) 298-312* 3.6 348-362* 3.1 408-462 19.2 478-502 5.6 533-547 7.1 643-657 3.8 673-702 12.5 713-737 3.7 873-892 4.4 898-917 6.6 943-957 5.5 968-982 17.5 1058-1092 7.3 1158-1192 3.8 1198-1252 8.8 1263-1277 4.9 1433-1452 5.6 1478-1492 7.9 1983-2016 5.5 14 Online References 1. Goser S, Andrassy M, Buss SJ, Leuschner F, Volz CH, et al. (2006) Cardiac troponin I but not cardiac troponin T induces severe autoimmune inflammation in the myocardium. Circulation 114: 16931702. 2. Kmecova J, Klimas J (2010) Heart rate correction of the QT duration in rats. Eur J Pharmacol 641: 187-192. 3. Radovits T, Korkmaz S, Loganathan S, Barnucz E, Bomicke T, et al. (2009) Comparative investigation of the left ventricular pressure-volume relationship in rat models of type 1 and type 2 diabetes mellitus. Am J Physiol Heart Circ Physiol 297: H125-133. 4. Pacher P, Nagayama T, Mukhopadhyay P, Batkai S, Kass DA (2008) Measurement of cardiac function using pressure-volume conductance catheter technique in mice and rats. Nat Protoc 3: 1422-1434. 5. Louch WE, Sheehan KA, Wolska BM (2011) Methods in cardiomyocyte isolation, culture, and gene transfer. J Mol Cell Cardiol 51: 288-298. 15 Online Figure 1. Online Figure 1 Both control and NaV1.5-peptide immunized rats exhibit premature ventricular beats. Representative surface 12-lead ECG tracing showing monomorphic premature ventricular beats (A; black arrows) from a NaV1.5-immnunized rat and ECG recordings by telemetry in a conscious NaV1.5-immnunized rat showing polymorphic premature ventricular beats (B; black arrows). 16 Online Figure 2 Immunization of rats with NaV1.5-peptide does not lead to myocardial inflammation and fibrosis. Histological examination of myocardium stained with hematoxylin and eosin (A) and Masson’s trichrome (B) in control and NaV1.5-immunized rats. Magnification x200, scale bar: 50µm 17 18 Online Figure 3 Immunization of rats with NaV1.5-peptide does not lead to myocardial inflammation and fibrosis. Histological examination of sinoatrial and atrioventricular node tissue with surrounding myocardium stained with hematoxylin and eosin and Masson’s trichrome in control rats and in NaV1.5-immunized rats. Circled areas indicate sinoatrial or atrioventricular nodes, black arrows indicate sinoatrial node artery. Magnification x40, scale bar: 200µm. 19 Online Figure 4 Immunization down-regulates myocardial NaV1.5 protein expression. Immunoblot analysis was performed for NaV1.5 protein band densities in ventricular myocardium using antibodies specific to NaV1.5 (Abcam, Cambridge, UK) in control (n=5) and NaV1.5-immunized (n=6) rats. 20 Online Figure 5 Sera from NaV1.5 immunized rats have no effect on voltage-gated calcium current (ICa) density in rat ventricular cardiomyocytes. (A) Representative recordings of ICa in cardiomyocytes after incubation with sera from control rats (left side) and with sera from Na V1.5 immunized rats (right side). (B) ICa densities after incubation with sera from controls (n=5 rats, 19 cardiomyocytes) or with sera from NaV1.5 immunized rats (n=4 rats, 19 cardiomyocytes). 21