Ginalski
advertisement

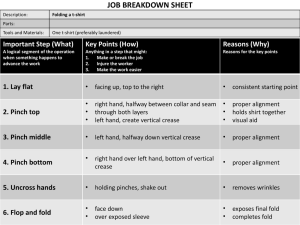
Ginalski (P0453) - 71 predictions: 71 3D structural and sequential constraints identified above. In some cases close homologues were also submitted to the Meta Server as the query sequences. For regions that displayed low stability (i.e. highly dependent on the server), possible alignment variants were derived manually, guided mainly by secondary structure predictions. Modeling of CASP5 Target Proteins with 3D-CAM K. Ginalski1,2 1 - Interdisciplinary Centre for Mathematical and Computational Modelling, Warsaw University, Warsaw, Poland, 2 - BioInfoBank Institute, Poznań, Poland kginal@bioinfo.pl All plausible alternative sequence-to-structure alignments were tested by building 3D molecular models for the target sequence with the Homology module of InsightII (Accelrys Inc., San Diego, CA). Backbone conformation was taken from the template structure, and only non-conserved side chains were substituted. Modeling of loops that contained insertion and deletion regions was skipped in this procedure. Models were then subjected to detailed evaluation, mainly by visual inspection of structural consistency and using Verify3D [5] and ProsaII [6] energy profiles. Such a 3D evaluation procedure enabled selection of final sequence-to-structure alignments. For the fifth round of Critical Assessment of Techniques for Protein Structure Prediction (CASP5), 67 target proteins were modeled using the 3D-Consensus Alignment Method (3D-CAM). The issue of sequence-to-structure alignment of target sequences with their respective parent structures was the main emphasis, and as shown in previous rounds of CASP, this part of the modeling procedure is the major source of errors. The critical steps in modeling: selection of template(s) and generation of sequence-to-structure alignment, were based on the results of secondary structure prediction and tertiary fold recognition carried out using the Meta Server [1]. Final models of target proteins were built using the MODELLER program [7]. Where possible, more than one template protein was used, after superimposition of their molecular structures. The overall quality of each modeled structure was checked in detail with the WHAT_CHECK program [8]. No energy minimization procedures were employed. Initially, related proteins with known structures were identified from the consensus of the Meta Server results. For difficult targets, template (fold) identification was based on the results of the 3D-Jury method (Rychlewski L., unpublished). Structural determinants of the fold were then analyzed: all the structures representing a given fold, and corresponding structural alignment extracted from the FSSP database [2], were inspected for both conservation and variability of the structural elements. Conservation of specific residues and contacts responsible for maintaining tertiary structure, and critical for substrate binding and/or catalysis, were also established. Additionally, homologous sequences that matched the targets were collected with PSI-BLAST searches [3] performed against the non-redundant protein sequence database and unfinished genomes until profile convergence. The CLUSTAL W program [4] was used to generate multiple sequence alignments for sets of sequences containing target, and other closely-related proteins, to identify conserved residues within the family. 1. 2. 3. 4. 5. 6. All alignments produced by different servers interacting with the Meta Server were inspected for both variability and violation of structural integrity. Initial alignment was obtained by taking, in most cases, the common alignment for each region (mainly for each secondary structure element), taking into account the structural alignment of templates where possible, within the context of the 7. 8. A-1 Bujnicki J.M. et al (2001) Structure prediction meta server, Bioinformatics 17 (8), 750-751. Holm L. et al, (1996) Mapping the protein universe, Science 273 (5275), 595-603. Altschul S.F. et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res, 25 (17), 3389-3402. Thompson J.D. et al (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice, Nucleic Acids Res, 22 (22), 4673-4680. Luthy R. et al (1992) Assessment of protein models with three-dimensional profiles, Nature 356 (6364), 83-85. Sippl M.J. (1993) Recognition of errors in three-dimensional structures of proteins, Proteins 17 (4), 355-362. Sali A. et al. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234 (3), 779-815. Hooft R.W. et al. (1996) Errors in protein structures. Nature 381 (6580), 272.