bit_22779_sm_SuppData
advertisement

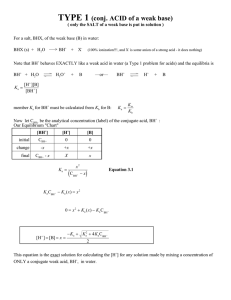
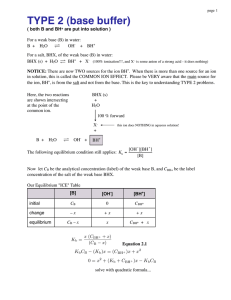
SUPPLEMENTARY ONLINE DATA Mechanism of rate retardation in cellobiohydrolase catalyzed cellulose hydrolysis Jürgen JALAK and Priit VÄLJAMÄE Institute of Molecular and Cell Biology, University of Tartu, Vanemuise 46 – 138, 51014 Tartu, Estonia Figure 1 (A) Hydrolysis of cellulose by CBH with (, , ) and without (, , ) the presence of 0.5 mM pNPL. and , 2.5 μM PcCel7D, PWS 1.0 g l-1; and , 2.5 μM TrCel7A, PWS 1.0 g l-1; and , 0.5 μM PcCel7D CD, PWS 2.5 g l-1. (B) Hydrolysis of 0.5 mM pNPL by 0.85 μM BG. This activity was not dependent on the type and concentration of cellulose present. Error bars are from 7 independent measurements. (C) Rate of the hydrolysis of pNPL by CBH in the presence of cellulose and BG at different concentrations. 0.5 mM pNPL, 2.5 μM TrCel7A, Avicel 10 g l-1. BG was , 0 μM; , 0.73 μM; , 0.85 μM; , 1.46 μM. (D) Hydrolysis of cellulose by CBH with (, ) and without (, ) the presence of 0.85 μM BG. and , 2.5 μM TrCel7A, Avicel 10 g l-1; and , 1.0 μM TrCel7A, PWS 10 g l-1. (E) All-free lines represent the hydrolysis of pNPL by CBH in the absence of cellulose. pNPL was 0.5 mM. In coordinates (vpNP/[CBH]) versus [pNP] data points obtained at CBH concentrations 0.5 - 2.5 μM fall in the same line. Solid lines represent non-linear regression according to the Equation (10). The values of kcat(pNPL), KM(pNPL) and Ki(Lac) of 5.25 min-1, 500 μM and 80 μM respectively were obtained for TrCel7A. Corresponding figures for PcCel7D were 4.0 1 min-1, 1000 μM and 36.7 μM. These figures were used in calculation of [CBH]FA from vpNP measured in the presence of cellulose according to the Equation (11) (see also supplementary Table I). , TrCel7A; , PcCel7D. (F) Binding kinetics of TrCel7A to Avicel. Total bound CBH ([CBH]bound, and ) was found from the binding experiment as difference between total CBH and CBH free from cellulose. CBH with occupied active site ([CBH]OA, and ) was found from parallel experiment according to the Equation (1). Avicel was 10 g l-1 and TrCel7A was 2.5 μM ( and ) or 1.0 μM ( and ). Table I. Calculation of observed catalytic constants (kobs)a for CBH catalyzed hydrolysis of cellulose Time min [pNP]b μM 0 0.5 2 5 10 15 20 30 40 60 0 1.65 5.75 12.22 21.10 28.65 35.40 48.14 59.31 78.12 vpNPc μM min-1 3.059 2.343 1.931 1.636 1.465 1.343 1.169 1.043 0.864 vpNP/[CBH]d min-1 2.59 2.52 2.41 2.26 2.15 2.06 1.89 1.76 1.57 [CBH]FAe μM 1.18 0.93 0.80 0.72 0.68 0.65 0.62 0.59 0.55 [CBH]OAf μM [Glc]g μM 1.32 1.57 1.70 1.78 1.82 1.85 1.88 1.91 1.95 0 20 65 128 209 274 331 434 525 688 vGlch μM min-1 kobs i min-1 35.3 23.9 18.0 14.3 12.4 11.2 9.6 8.7 7.5 13.42 7.63 5.32 4.03 3.41 3.03 2.56 2.28 1.93 experiment was conducted in 50 mM sodium acetate pH 5.0 at 25 ºC with magnetic stirring 500 rpm. Concentrations were following: [Avicel] = 10 g l-1, CBH was 2.5 μM TrCel7A, [pNPL] = 500 μM, [BG] = 0.85 μM. a b measured concentration of CBH released pNP. These figures were subjected to non-linear regression according to the Equation (6) in order to get parameter values for velocity calculations. c velocity of pNP formation calculated according to the Equation (8) using parameter values from previous step. d ratio vpNP/[CBH] was calculated from all-free line for TrCel7A according to the Equation (10) using measured values for [pNP]b. These figures represents the expected values of (vpNP/[CBH]) at each [pNP]b without the presence of cellulose. e concentration of CBH with free active site for pNPL hydrolysis is calculated from the figures in previous two columns. [CBH]FA = vpNP/(vpNP/[CBH]) (Eq. 11). f concentration of CBH with occupied active site is the difference between total CBH ([CBH]Tot) and [CBH]FA. In this example [CBH]OA = 2.5 μM - [CBH]FA. g measured concentration of CBH released glucose equivalents. These figures were subjected to nonlinear regression according to the Equation (7) in order to get parameter values for velocity calculations. h velocity of glucose formation calculated according to the Equation (9) using parameter values from previous step. 2 i observed catalytic constant for cellobiose formation is now available from kobs = vGlc/2[CBH]OA (Eq. 2, velocity of cellobiose formation was considered to be equal to the half of the measured rate of glucose formation in the presence of BG). Table II Relative observed catalytic constants of TrCel7A, PcCel7D and their catalytic domains (CD-s) on different celluloses Relative kobs Cellulose Cel7A Cel7D Cel7A CD Cel7D CD 100 46 ± 5 48 ± 4 44 ± 11 100 49 ± 6 117 ±26 60 ± 9 a BMCC Avicel RAC PWS 100 48 ± 5 57 ± 8 42 ± 8 % 100 43 ± 2 72 ± 5 41 ± 14 a Relative kobs (as % of the value on BMCC) were first taken for each time point separately and table lists the average values over all time points. Table III Observed catalytic constants for catalytic domains (CD-s) compared to that of intact enzymes Observed catalytic constants for TrCel7A and its CD compared to corresponding figures for PcCel7D Relative kobs Cellulose Cel 7 ACD Cel 7 A Cel 7 DCD Cel 7 D %a Cel 7 A Cel 7 D Cel 7 ACD Cel 7 DCD BMCC Avicel RAC PWS 79 ±10b 76 ± 5 85 ± 8 81 ± 6 53 ± 7b 60 ± 7 83 ± 7 81 ± 12 56 ± 8c 62 ± 4 43 ± 3 58 ± 5 84 ±18d 78 ± 7 45 ± 11 58 ± 5 a Relative kobs were first taken for each time point separately and table lists the average values over all time points. b Relative kobs for CD as the % of the corresponding value for intact enzyme. c Relative kobs for TrCel7A as the % of the corresponding value for PcCel7D. d Relative kobs for TrCel7A CD as the % of the corresponding value for PcCel7D CD. 3