Campbell Mutant Protocol
advertisement

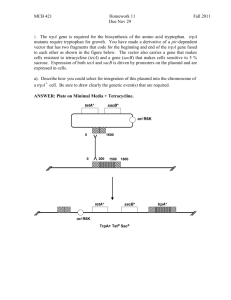
V. fischeri Campbell Mutants This is a basic protocol for the construction of V. fischeri Campbell mutants. Essentially, the procedure works by crossing a plasmid (which contains a short stretch of homologues sequence from the gene of interest) into the V. fischeri chromosome. The gene is inactivated by the insertion of the plasmid sequence (which should include a selectable marker) in the middle of the gene's coding sequence. The advantages: Since this method involves only a single cross-over, it's much faster than constructing a true null mutant by marker exchange. Only one selectable marker is used (the one on the vector), so working out a scheme of antibiotic selection is easier as well. The disadvantages: No sequence is actually deleted from the gene, so the chromosome retains all the coding sequence necessary for the functional gene. This means that, should the plasmid cross back out, the strain effectively reverts to wild-type. For this reason, selection (i.e. antibiotic exposure) should be maintained on these mutants whenever possible (the Visick lab has reported very low reversion rates during animal experiments w/out antibiotics though). The protocol: 1. Design primers to amplify a fragment from the gene of interest. We generally shoot for ~150 bps, but the Visick lab has made campbells with as few as 80 bp fragments. Remember that the crossover will occur at the site of the fragment, so try to target the front half of the gene. Also, consider building restriction sites into your primers (these will be selected based on the plasmid that you'll be ligating the fragment into) and don't forget to add an extra GC to the ends the restriction enzyme target sequence (this gives the enzyme a bit of "scaffolding" to sit down on). 2. PCR amplify the gene fragment from a genomic DNA prep (usually from ES114). Confirm that the amplimer is of the appropriate size, and gel purify. 3. In separate reactions, digest the amplimer and a prep of the chosen plasmid with the appropriate restriction enzyme(s). Run the resultant digests through the QiaQuick PCR purification kit to remove the oligos trimmed off the ends (this step also removes the enzyme). 4. Ligate the cut amplimer into the cut plasmid using T4 DNA ligase. See the product sheet for appropriate DNA concentrations, etc. We generally do overnight ligations with alternating cycles of 10 and 30°C (program file 61 in the PE Cetus thermocycler). 5. Transform an appropriate E. coli strain using the ligation mix (i.e. your putative plasmid construct) to transform. For most of the R6K origin plasmids we use around the lab, the strain to use is usually DH5α λpir (pre-prepped competent Prepared by Joe Graber, June 2005 cells are usually available; check in the -80°). Plate the transformed coli strain on media (LB or BHI) containing the appropriate antibiotic (i.e. whatever's on the plasmid). 6. The next morning, check the plates for colonies. If you have one or more, pick it/them and, a) make freezer stocks, b) set up overnight cultures for a plasmid prep. Don't forget to maintain antibiotic selection throughout (otherwise, the strains are likely to lose the plasmids) 7. Using the plasmid prep, confirm construction of the desired vector. You should use two methods to do this: - Use your original primers in a PCR amplification, using the construct as a template and the original plasmid as a control; confirm that you have single amplimer of the expected size. - Digest your plasmid with at least two restriction enzyme, including on that cuts within the insert; confirm that all fragments (and the additive total of the fragments) are of the expected size. 8. Now that you've confirmed your plasmid construct, it's time to move it into V. fischeri. Do a tri-parental mating (see the appropriate protocol) or whatever other method you're partial to. Plate on LBS media containing the appropriate antibiotic, and pick resistant colonies. Immediately make freezer stocks. 9. Use two methods to confirm the mutant: - Using genomic DNA as a template (and ES114 gDNA as a control), PCR amplify with primers targeting sequences a few hundred bps up and downstream of the putative insertion site. Obviously, you should see a much larger amplimer from the mutant strain. - Do a Southern (see the appropriate protocol) with probe targeting the antibiotic cassette to confirm a single chromosomal insertion of the plasmid. If all goes as planned, you should have your mutant. Now, go do some science. Prepared by Joe Graber, June 2005