3 Physiological Ecology: Resource Exploitation by Microorganisms 52

3 Physiological Ecology: Resource Use by Microorganisms
Outline:
3.1 Diverse habitats provide selective pressures over evolutionary time and cause physiological diversity.
3.2 Genomics can help us understand biology and evolution
3.3 Organisms use carbon- and energy-sources, so make physiological ecology.
3.4 Ecosystem nutrient changes regulate what is in, and the physiological state of microbial communities. This results in selective pressures.
3.5 Cellular responses to starvation include resting stages, environmental sensing, gene regulation, dormancy, and slow growth.
3.6 A surface of the planet has complex mixtures in chemical disequilibrium.
3.7 A thermodynamic hierarchy can describe biosphere selective pressures, energy sources, and biogeochemical reactions.
3.8 The thermodynamic hierarchy of half reactions can be used to predict biogeochemical reactions in time and space.
3.9 Through an overview of metabolism we can see the “logic of electron transport”.
310 The flow of carbon and electrons in anaerobic food chains show that syntrophy is most important.
3.11 There is a large diversity of lithotrophic reactions.
3.1 Diverse habitats provide selective pressures over evolutionary time and cause physiological diversity.
Metabolism, replication, and heredity are part of life. These three processes work in the environment that provides resources for metabolism and selective pressures for both replication and heredity. There is a long (3.8 x 10 9 years) and active interaction between
Earth’s habitats and microorganisms. By examining thermodynamically favored geochemical reactions, we can understand and predict selective pressures that act on microorganisms.
There are links between the planet’s habitat diversity and what is in the genomes of microorganisms. Diversity in habitats means there is diversity in selective pressures and resources. Through evolution, the result is molecular, metabolic, and physiological diversity in microorganisms.
3.2 Genomics give biological and evolutionary insights
Genome size
Through studying genomics we can begin to understand what prokaryotes are and their evolutionary responses to past environments. The sequence of an organism’s genome shows what it can do now and its historical physiological capabilities. In the last decade, genomesequencing has provided a large amount of basic information about microbial life. Over 450 bacterial genomes, 35 archaeal genomes, and 15 eukaryotic microbial (fungi, protozoa)
1
genomes were either in completed or being found. Hundreds more are being determined.
These prokaryote genomes come from ocean water, soils, sediments, sewage, or hot springs.
These are microorganisms whose physiological properties make them valuable for understanding environmentally significant biogeochemical processes such as the cycling of carbon, nitrogen, sulfur, metals, and organic environmental pollutants.
There is large variation in size (number of amino acids) among proteins, but they average somewhat more than 300 amino acids each. At three codons per amino acid, each gene consists of about 10 3 bases. Bioinformatic software divides the long stretches of DNA bases into ORFs based on recognition of start and stop sites for DNA replication and protein translation on the encoded mRNA.
Two strains of the same species can possess very different genotypes and phenotypes. This is shown by Escherichia coli and Prochlorococcus. Gaining DNA by an ancestor of an E. coli strain appears to have changed the gentle intestinal bacterium to an often lethal human pathogen.
Similarly, the Prochlorococcus strain with the larger genome has been shown to live in an ecological niche characterized by low light intensity.
There is a wide range in sizes of prokaryotic genomes. The largest prokaryotic genomes are free-living heterotrophic Bacteria from the soil. It is argued that the large genomes of soildwelling microorganisms have many genes (both regulatory and structural) that give qualities such as slow growth and exploitation of resources that are both diverse and scarce.
Gene content within microbial genomes
After finding individual genes (ORFs), then the ORFs are aligned with sequences of homologous genes of known function. These two steps allow amino acid similarities in proteins encoded by new and previously characterized genes to be computed so that the biochemical function of the new genes can be determined.
We don’t completely understand prokaryotic genomes and prokaryotic biology. The proportion of unknown hypothetical genes in all genome-sequencing projects is roughly onethird, even for the thoroughly studied bacterium, E. coli. Maybe poorly characterized genes have no function, but maybe these genes show how little we understand about real-world ecological pressures and the physiological processes expressed by microorganisms in their native habitats. Because laboratory cultivation of microorganisms is very different from conditions experienced by microorganisms in the field, many genetic traits may go
unexpressed by the organism, so they are unobserved by the microbiologist. Understanding the function of unknown hypothetical genes represents one of the major frontiers in biology.
Integrating genome data
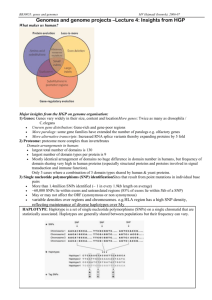
What we can learn from the portion of each genome that we understand is useful. For each sequenced genome, a template for cell function is used to organize the genes. Figure 3.2 shows the metabolic template for a model cell. A generalized cell model has an interior
2
(cytoplasm) and an exterior bounded by the cytoplasmic membrane and the cell wall. The outside edge of the cell is lined with membrane-bound transport proteins that regulate cytoplasmic composition by acting on inorganic cations, inorganic anions, carbohydrates, amino acids, peptides, purines, pyrimidines, other nitrogenous compounds, carboxylates, aromatic compounds, other carbon compounds, and water. Many of the transport mechanisms are energy-coupled (requiring ATP) and show a recognizable ATP binding cassette (ABC) motif. Channel proteins transport materials both into and out of the cell, as do
P-type ATPase transport systems for input and output.
The inside of the model cell contains a template for organizing products (proteins) of recognized genes involved in metabolism. Similar templates apply to genes functioning in information storage and other cellular processing. Structural and regulatory genes encoding metabolic processes (respiration, the tricarboxylic acid cycle, other biochemical pathways, etc.) appear as networks within the cell.
3
Figure 3.3 is an overview of the integrated genetic information for metabolism and transport in Pseudomonas putida. Analysis of this 6.18 Mb genome provides a model for the physiological and ecological function of pseudomonads – a broad class of opportunistic and adaptable bacteria that are widespread in terrestrial and aquatic environments. The genome data are the basis for many hypotheses about genetic determinants for transporters, oxygenase enzymes, electron transport chains, sulfur metabolism proteins, and microbial mechanisms for protection from toxic pollutants and their metabolites.
Figure 3.3
Genomic representations such as that shown in Figure 3.3 provide good ideas of what microorganisms are, what they can do, and what each of their evolutionary histories have been. Genome sequencing extends our understanding of prokaryotic biology. To test hypotheses that arise from genome sequencing data, other essential tools include biochemical assays, physiological assays, transcriptome analysis, proteome analysis, sitedirected mutagenesis creating knockout and other mutations, genetic complementation, computational modeling, systems biology , and tests of ecological relevance.
4
Here is a generalized view of metabolism.
Enzymes direct the reactions in biochemical pathways
5
Enzymes work by making reactions go faster.
3.3 Organisms use carbon- and energy-sources. These uses are a foundation for physiological ecology.
All forms of life (from prokaryotes to humans) need to live. They all need at least two physiological resources: (i) an energy source for generating ATP; and (ii) a carbon source for assembling the cellular building blocks during maintenance of existing cells and/or creation of new cells (growth). No life form can exist without using well-defined energy and carbon sources. Therefore, when microbial physiologists and microbial ecologists study a new organism or a new habitat, the first questions to ask are: “What drives metabolism here?
What are the energy and carbon sources?” The answers provide fundamental nutritional bases for later hypotheses and inquiries about past and ongoing biogeochemical and ecological processes.
Table 3.3 provides a useful way of classifying the nutritional needs of both individual microorganisms and ecosystems. The matrix shows the energy source along the top and carbon source along the left side. Energy sources fall into two main categories: chemical and light. The chemical sources of energy are further divided into two: inorganic (such as hydrogen gas, ammonia, methane, and elemental sulfur) or organic compounds (generally containing multiple C-C bonds). The carbon sources also fall into two categories: gaseous CO
2 or fixed organic carbon. The matrix in Table 3.3 provides a way to nutritionally classify nearly all forms of life on Earth into five groups. The two that are perhaps easiest to recognize are plants and animals (including humans). Plants are photosynthetic autotrophs – getting energy from light and carbon from gaseous CO
2
. Humans, fungi, and microorganisms that cycle plantderived (and other) organic substrates are chemosynthetic organoheterotrophs.
Microorganisms that oxidize inorganic compounds (e.g., hydrogen gas, ammonia, elemental sulfur) for energy and bring CO
2
into their biomass are chemosynthetic lithoautotrophs.
Microorganisms that use light as an energy source and bring fixed carbon into their biomass are photosynthetic heterotrophs (this lifestyle is exclusively prokaryotic and rare).
6
Microorganisms that use inorganic compounds as energy sources and bring fixed carbon into their biomass are chemosynthetic lithoheterotrophs (this lifestyle is exclusively prokaryotic and extremely rare). It should be recognized that the boundaries between the five lifestyle categories in Table 3.3 may sometimes be blurred. For instance, some microorganisms are genetically and physiologically versatile – adopting a heterotrophic lifestyle when fixed organic carbon is available, then resorting to CO
2
fixation in the absence of organic carbon.
COG analysis of three organisms in each nutritional class suggests that the transport and metabolism of both amino acid and carbohydrates have been emphasized during the evolution of chemosynthetic organoheterotrophs (especially soil-dwelling bacteria). For photosynthetic heterotrophs, a relatively large portion of the genome has been for energy production and energy conservation. Also, there seems to be a trend in photoautotroph evolution that reduces the need for inorganic ion transport and metabolism. The idea of links between evolutionary pressures and gene content is useful.
7
Figure 3.4 Metabolic genes (as percent of total genome) in representatives of the four major nutritional categories in prokaryotes: chemosynthetic organoheterotrophs (CSH), photosynthetic heterorophs (PSH), chemosynthetic lithoautotrophs (CSLA), and photosynthetic autotrophs (PSA). Bar graph shows the percent of the genome coding for amino acid transport + metabolism, carbohydrate (CH2O) transport + metabolism, energy production + conversion, and inorganic ion transport + metabolism. Data are based on COG analysis of 12 genomes.
Glossary anaerobic metabolism ,
혐기성 대사
[
嫌氣性代謝
]
분자상 산소의 소비를 동반하지 않는 에너지대사
.
산화환원반응도 포함되지만 산소를
최종 전자수용체로 사용하지 않기 때문에
,
대신유기중간체를 환원하여 소위 발효산물을
만들고
CO2
로의 완전산화는 되지 않는다
.
대표적인 것은 해당 및 각종 발효과정이다
.
이때 탄수화물 등의 분해과정에서 중간에 생기는 고에너지 인산화합물로부터
ATP
를
형성하지만 호기적 대사보다 능률은 훨씬 나쁘다
. cellular ,
세포의
8
[
細胞
]
세포라는 영어단어
cell
의 형용사형
. disequilibrium ,
비평형
현실의 어떤 면들이 개인의 지식 구조
(
도식
)
와 맞지 않을 때 생기는
‘
불편한
’
상태
. dormancy ,
휴면
[
休眠
]
생물의 활동 또는 생장이 일시적으로 정체되거나 정지되는 일을 말하는데 겨울철의
휴면을 동면
(
冬眠
:
겨울잠
),
여름철의 휴면을 하면
(
夏眠
)
이라고 한다
.
동물과 식물에서
모두 볼 수 있다
. gene regulation ,
유전자 조절
[
遺傳子調節
]
Controlling how much, how fast, and what type of protein is made in a cell. genome ,
게놈
한 생물이 가지는 모든 유전 정보를 말하며 유전체라고도 한다
.
일부 바이러스의
RNA
를
제외하고 모든 생물은
DNA
로 유전 정보를 구성하고 있기 때문에 일반적으로
DNA
로
구성된 유전 정보를 지칭한다
. genomics ,
유전체학
[
遺傳體學
]
생물의 디옥시리보 핵산
(DNA),
리보 핵산
(RNA)
과 같은 유전 정보를 밝히고자
유전체
(genome)
를 단위로 실험 구상과 정보 처리를 수행하는 학문
.
유전체학의 가장
기본 단계는 인간을 포함한 각종 생물들의 유전체 염기 서열 판독
(sequencing)
으로서
주로 알려진 서열에서 염색체 지도와 유전자 지도를 비교 분석하여
, DNA
구조 결정 등을
연구한다
. 1990
년대초 미국에서 시작된 유전체 프로젝트
(genome project)
의 추진에 따라
탄생한 학문이다
. genotype ,
유전자형
[
遺傳子型
]
9
생물이 가지고 있는 특정한 유전자의 조합
.
또는 그 생물 개체가 가지고 있는 유전자
전체의 집합을 가리키기도 한다
. half reaction ,
산화환원짝
산화
-
환원짝은 산화 환원 반응에서 산화되는 반쪽 반응과 환원되는 반쪽 반응을 일컫는
말이다
. heterotrophic ,
종속영양의
[
從屬營養
~ ]
탄소원
(
炭素源
)
으로서 유기물을 사용하는 영양 형태로
,
독립영양에 대응하는 것으로
종속영양생물서 유기물은 몸체의 구성물의 원료로 필요할 뿐만 아니라 혐기적
(
嫌氣的
)
분해
(
해당
,
발효
)
혹은 호기적
(
好氣的
)
분배
(
호흡
)
에 의하여 에너지를 취하는 원
(source)
이
됨
.
동물
,
녹색이 아닌 식물세포 및 대다수의 미생물은 이 형에 속하며 식물도 어두운
곳에서는 종속영양의 방식을 취함
.
즉
,
종속영양은 생육을 위하여 필요한 영양원을 이미
만들어진 유기물에 의존하는 영양 형식이며
,
무기물로부터 그것을 합성한
독립영양
(
獨立營養
)
과 대응되는 생물의
2
대 영양의 하나임
.
ORF, open reading frame,
개방형 해독틀
아미노산의 서열로 번역되는
DNA
염기서열을 가리키는 말이다
.
즉 번역개시코돈
(start codon; ATG)
에서 정지코돈
(stop codon; TGA, TAA, TAG)
에 이르는 염기서열을 말한다
. parasite ,
기생충
어떤 생물
(
기생체
)
이 인간 또는 가축 등
(
숙주
host)
의 체표 또는 체내에 머물면서 더구나
영양을 얻고 숙주에게는 해를 미치게 하는 상태를 기생생활
(parasitism)
이라고 한다
. phenotype ,
표현형
[
表現型
]
생물에서 겉으로 드러나는 여러 가지 특성
.
물리적인 특성뿐만 아니라 행동 같은
특성까지도 포함한다
. selective pressure ,
선택압
[
選擇壓
]
10
자연돌연변이체를 포함하는 개체군에 작용하여 경합에 유리한 형질을 갖는 개체군의
선택적 증식을 재촉하는 생물적
,
화학적 또는 물리적 요인
.
세균집단은 일반적으로
10–
8~10-7
의 비율
(
돌연변이율
)
로 자연돌연변이주가 생겨 여러 가지 비율
(
돌연변이빈도
)
로
개체군 내에 혼재한다
.
이들은 정상 환경조건에서는 분열속도가 변화하지 않는 한
,
이
집단에 있어서는 변화하는 일은 없다
.
이러한 집단에 대해 변이주에 있어 유리한
환경요인이 작용하면 변이주의 급속한 선택적 증식이 일어나 집단변이로 이어진다
.
이것을 도태라고 하고 그 요인을 도태압이라고 하기도 한다 transcriptome analysis ,
전사체 해석
[
轉寫體解析
]
전사체는전사와 유전체 합성체로
,
체세포에서의 전사체의 유전자 발현량을 규명하는 것
.
암처럼 표적으로 삼는 조직
(
또는 세포
)
이뚜렷한 것을 대상으로 하며
,
주로
DNA
칩을
사용한다
. DNA
칩에 많은
(
수만 종
) cDNA
를 표적하여
,
여기에 다른 세포 또는다른 조건
하에서 채취하여 각각 다른 색으로 표지한
cDNA
를놓고 잡종교잡을 형성하면서 다수의
유전자 발현량
(
발현엽록소
)
을 조사한다
.
이 방법으로 세포가 놓여진 상태의 차이를
각유전자 차이의 총화로 파악할 수 있다
.
이로써 각각의 암세포성질에 따라 주문제
치료를 실현할 수 있게 된다
.
11