Feature weighting for nearest neighbor algorithm by Bayesian
advertisement

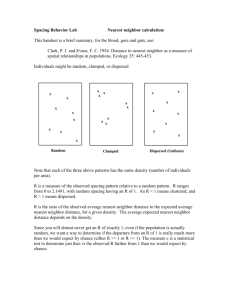
FEATURE WEIGHTING FOR NEAREST NEIGHBOR ALGORITHM BY BAYESIAN NETWORKS BASED COMBINATORIAL OPTIMIZATION Iñaki Inza (ccbincai@si.ehu.es), post-graduate student. Dpt. of Computer Sciences and Artificial Intelligence. Univ. of the Basque Country. Spain. Abstract - A new approach to determining feature weights for nearest neighbor classification is explored. A new method, called kNN-FW-EBNA (Nearest Neighbor Feature Weighting by Estimating Bayesian Network Algorithm), based on the evolution of a population of a discrete set of weights ( similar to Genetic Algorithms [2] ) by EDA approach ( Estimation of Distribution Algorithms ) is presented. Feature Weighting and EDA concepts are briefly presented. Then, kNN-FW-EBNA is described and tested on Waveform-21 task, comparing it with the classic K-NN unweighted version. Nearest neighbor basic approach involves the storing of training cases and then, when given a test instance, finding the training cases nearest to that test instance and using them to predict the class. Dissimilarities among values of the same feature are computed and added, obtaining a representative value of the dissimilarity between compared pair of instances. In basic nearest neighbor approach, dissimilarities in each feature are added in a ‘naive’ manner, weighing dissimilarities equally in each dimension. This approach is handicapped, allowing redundant, irrelevant, and other imperfect features to influence distance computation. However, when features with different degrees of relevance are present, the approach is far from the bias of the task. See Wettschereck et al. [13] for a review of feature weighting methods for nearest neighbor algorithms. A search engine, EBNA [5], based on Bayesian networks and EDA [11] paradigm, is the basis to state the algorithm within search parameters: searching for a set of discrete weights for the nearest neighbor algorithm. Evolutionary computation groups a set of techniques – genetic algorithms, evolutionary programming, genetic programming and evolutionary strategies - inspired on the model of organic evolution, which constitute probabilistic algorithms for optimization and search. In these algorithms, the search in the space of solutions is carried out by means of a population of individuals – possible solutions to the problem - which, as the algorithm is developed, evolves through more promising zones of the search space. Each of the previous techniques requires the design of crossover and mutation operators in order to generate the individuals of the next population. The manner in which individuals are represented is also important, because depending on this and on the former operators, the algorithm will take into account, in one implicit way, the interrelations between the different pieces of information used to represent the individuals. An attempt to design evolutionary algorithms for optimization based on populations that avoid the necessity to define crossover and mutation operators specific to the problem, and which are also able to take into account the interrelations between the variables needed to represent the individuals, is the so called Estimation of Distribution Algorithms (EDA). In EDA there are no crossover nor mutation operators, the new population is sampled from a probability distribution which is estimated from the selected individuals. This is the basic scheme of EDA approach: Do Generate N individuals ( the initial population ) randomly. Repeat for l = 1,2,... until a stop criterion is met. DSl-1 Select S <= N individuals from Dl-1 according to a selection method. pl(x) = p(x | DSl-1 ) Estimate the probability distribution of an individual being among the selected individuals. Dl Sample N individuals ( the new population ) from pl(x). The fundamental problem with EDA is the estimation of pl(x) distribution. One attractive possibility is to represent the n-dimensional pl(x) distribution by means of the factorization provided by a Bayesian network model learned from the dataset constituted by the selected individuals. Etxeberria and Larrañaga [5] have developed an approach that uses the BIC metric ( Schwarz [12] ) to evaluate the fitness of each Bayesian network structure in conjunction with a greedy search ( Buntine [4] ) to obtain the first model. The following structures are obtained by means of a local search that starts in the model found in the previous generation: this approach was called EBNA ( Estimating Bayesian Network Algorithm ). We will use this EBNA approach as the motor of the search engine that seeks for an appropriate set of discrete feature weights for the nearest neighbor algorithm. After Feature Weighting ( FW ) problem and EBNA algorithm are presented, the kNN-FW-EBNA approach can be explained: kNN-FW-EBNA is a feature weight search engine based on the ‘wrapper idea’ [7]: the search is guided through the space of discrete weights using 5 fold – cross validation error of the nearest neighbor algorithm in training set as evaluation function. In order to learn the n-dimensional distribution of selected individuals by means of Bayesian networks, populations of 1,000 individuals are used. Half of the best individuals, based on the value of the evaluation function, are used to induce the Bayesian network that estimates pl(x). The best solution found through the overall search is presented as the final solution when the next stopping criterion is reached: the search stops when in a sampled new generation of 1,000 individuals no individual is found with an evaluation function value improving the best individual found in the previous generation. Do Generate 1,000 individuals randomly and compute their 5 fold – cross validation error. Repeat for l = 1,2,... until the best individual of the previous generation is not improved. DSl-1 Select the best 500 individuals from Dl-1 according to the evaluation function. BNl (x) induce a Bayesian network from the selected individuals. Dl Sample by PLS [6] 1,000 individuals ( the new population ) from BNl (x) and compute their 5 fold – cross validation error. Experiments with 3 (0,0.5,1),5 (0,0.25,0.5,0.75,1) and 11 (0,0.1,0.2,...,0.9,1) possible weights were run on the Waveform-21 task ( see Breiman et al. [3] for more details ): a 3 class and 21 feature task with features with different degrees of relevance to define the target concept. In the learned Bayesian network each feature of the task was represented by a node, its possible values being the set of discrete weights used by the nearest neighbor algorithm. Except for the weight concept, dissimilarity function presented in Aha et al. [1] was utilized, using only the nearest neighbor to classify a test instance (1-NN). 10 trials with different training and test sets of 1,000 instances were created to soften the random starting nature of kNN-FW-EBNA and reduce the statistical variance. This sample size was vital to work with a standard deviation of the evaluation function lower than 1.0 %: higher levels of uncertainty in the estimation of the error can overfit the search and evaluation process, obtaining high differences between the estimation of the error in the training set and the generalization error on unseen test instances. We hypothesized that the unweighted nearest neighbor algorithm would be outperformed by kNN-FW-EBNA in each set of possible weights. Average results on test instances are reported in Table 1, instances which do not participate in the search process. Accuracy level CPU time Stopped generations Size of search space Unweighted approach 76.70 % 0.30 % 30.56 -------3 possible weights 3,3,3,3,4,2,2,2,3,3 321 77.72 % 0.37 % 85,568.0 5 possible weights 4,4,3,3,3,4,3,3,4,3 521 77.85 % 0.50 % 103,904.0 11 possible weights 77.76 % 0.54 % 116,128.0 4,4,4,4,4,4,3,3,4,4 1121 Table 1: average accuracy levels, standard deviations and average running times in seconds for a SUN SPARC machine from 10 trials for Waveform-21 task are presented. Generation where each trial stopped ( initial population was generation ‘0’ ) and search space cardinalities are also presented. A t-test was run to see the degree of significance of accuracy differences between the found discrete set of weights and the unweighted approach: thus, differences were always significant in a = 0.01 significance level. On the other hand, differences between feature weighting approaches were never statistically significant: a bias-variance trade-off analysis of the error [9] respect the number of possible discrete weights (3,5 or 11) which will give us a better understanding of the problem. High cost of kNN-FW-EBNA must be marked, increased with the size of the search space. The work done by Kelly and Davis [8] with genetic algorithms and that done by Kohavi et al. [10] with a best-first search engine to find a set of weights for the nearest neighbor algorithm can be placed near kNN-FW-EBNA. This work can be seen as an attempt to join two different worlds: on the one hand, Machine Learning and its nearest neighbor paradigm and on the other, the world of the Uncertainty and its probability distribution concept: a meeting point where these two worlds collaborate with each other to solve a problem. [1] D. W. Aha, D. Kibler and M. K. Albert. Instance-based learning algorithms. Machine Learning, 6, 3766, 1991. [2] Th. Bäck. Evolutionary Algorithms in Theory and Practice. Oxford University Press. 1996. [3] L. Breiman, J. H. Friedmann, R. A. Olshen and C. J. Stone. Classification and Regresion Trees. Wadsworth & Brooks, 1984. [4] W. Buntine. Theory refinement in Bayesian networks. In Proceedings of the Seventh Conference on Uncertainty in Artificial Intelligence, pages 102-109, Seattle, WA, 1994. [5] R. Etxeberria and P. Larrañaga. Global Optimization with Bayesian networks. II Symposium on Artificial Intelligence. CIMAF99. Special Session on Distributions and Evolutionary Optimization, 1999. [6] M. Henrion. Propagating uncertainty in Bayesian networks by probabilistic logic sampling. In Proceedings of the Fourth Conference on Uncertainty in Artificial Intelligence, pages 149-163, 1988. [7] G. John, R. Kohavi and K. Pfleger. Irrelevant features and the subset selection problem. In Machine Learning: Proceedings of the Eleventh International Conference, pages 121-129. Morgan Kaufmann, 1994. [8] J. D. Kelly and L. Davis. A hybrid genetic algorithm for classification. Proceedings of the Twelfth International Joint Conference on Artificial Intelligence, pages 645-650. Sidney, Australia. Morgan Kaufmann, 1991. [9] R. Kohavi and D. H. Wolpert. Bias plus variance decomposition for zero-one loss functions. In Lorenzo Saitta, editor, Machine Learning: Proceedings of the Thirteenth International Conference. Morgan Kaufmann, 1996. [10] R. Kohavi, P. Langley and Y. Yun. The Utility of Feature Weighting in Nearest-Neighbor Algorithms. ECML97, poster, 1997. [11] H. Mühlenbein, T. Mahnig and A. Ochoa. Schemata, distributions and graphical models in evolutionary optimization, 1998. Submitted for publication. [12] G. Schwarz. Estimating the dimension of a model. Annals of Statistics, 7(2), 461-464, 1978. [13] D. Wettschereck, D. W. Aha and T. Mohri. A Review and Empirical Evaluation of Feature Weighting Methods for a Class of Lazy Learning Algorithms. Artificial Intelligence Review, 11, 273-314, 1997.