Expt 1 Bacteria Harvest and Solution Preparation
advertisement

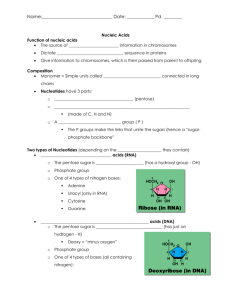
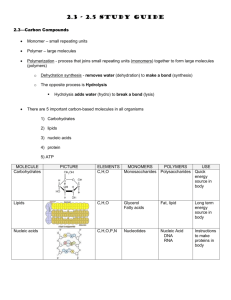
Chem 152 : Biochemistry laboratory Department of Chemistry School of Science and Engineering Loyola Schools, Ateneo de Manila University Nucleic Acids 1- Microbiology and Preparation Gloves are needed for all lab days dedicated to nucleic acids. Introduction Nucleic acids are a family of biopolymers that carry genetic information universally found in all living things from viruses and bacteria, to human cells, and are composed of chains of monomers of nucleotides. Each nucleotide consists of three parts: a nitrogenous base that is either a purine or a pyrimidine that project from a 5’ carbon pentose sugar backbone, and a phosphate group. The most famous nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). DNA molecules are known as the largest biologically active molecule with MW > 1 x 108 Daltons. Johann Friedrich Miescher discovered nucleic acids in 1968, naming them “nuclein” since the material was found first found in the nucleus. Prokaryotes, which do not contain a nucleus, were later also known to possess nucleic acids. (From NIMBB’s Dr. Cynthia PalmesSaloma, Ph.D.’s introduction to Nucleic Acids paper, CHED Faculty Upgrading Workshop in Molecular Biochemistry 2007). For today’s experiment, you will be preparing and growing bacterial cultures from which you will select a colony to propagate for DNA sample purification in the next few weeks. For this, you will need to learn how to operate the autoclave and work using some basic sterile techniques in microbiology. General Notes - For the nucleic acid set, use only autoclaved glassware, microcentrifuge tubes, pipet tips, and solutions. - Work division: o 26 November : Bacteria harvest, preparation of solutions for next week Materials autoclaving for bacteria inoculation on November 24 (c/o instructor) Bacteria inoculation on November 25 (c/o students) o 3 December : Extraction of samples (E.coli, banana, onion, liver) Materials and solutions autoclaving on 2 December o 10 December: DNA gel run and staining Prelab - Research on bacterial plate streaking and commercial/common bacterial growth media (at least two, one is LB). Procedure Nov. 24 Instructor will do this. Prepare the following for autoclaving: a. (7) 250-mL Erlenmeyer flasks with 50 mL LB (liquid medium) each. Concentration for all LB liquid medium is 20 g/L. b. Flask with 120 mL LB agar (32 g/L) c. Toothpicks d. Inoculation rods e. Petri dishes (7) f. A beaker full of microcentrifuge tubes g. Blue and yellow pipetor tips h. 1-mL 70% glycerol solution - Note that all flasks must have cotton plugs and must be covered with aluminum foil at the top. Student will do this.(Nov. 25) Bacterial inoculation: 1. Before doing the inoculation process, the student must observe proper sterilization techniques. Student must put on mask and gloves, and wear apron. Before using the safety cabinet, make sure the equipment is sterilized by spraying 70% ethanol solution on the surface. Using tissue paper wipe off the sprayed solution to dry the surface. 2. Each group will be given one flask containing LB broth. 3. Using a sterilized inoculation loop/rod, select and transfer one bacteria colony to the LB broth. Students must observe proper sterilization techniques while dealing with bacteria culture. 4. Cover the flask with the cotton plug and foil. 5. Put the flask with the culture in the shaking water bath which is already set to 37ºC and with 110 rpm shaking. 6. Leave the flask in the shaking water bath overnight. Nov. 26 1. Bacteria Harvest: a. Turn off the shaking water bath and remove the flask from the bath holder. b. Transfer ~ 8 mL of the culture to the centrifuge tube provided. c. Put the tube in the refrigerated centrifuge and run the equipment using these parameters: 10000 rpm, for 5 minutes. d. After the run, discard the supernatant to the waste container. e. Repeat steps b to d until all the broth have been centrifuged. f. Store the harvested bacteria in the refrigerator for processing next week. 2. Prepare the following solutions for next week’s extraction: a. About 100 mL of 1% SDS solution b. 1 M Tris-HCl c. d. e. i. Measure 18.17 g of the solid Tris, then dilute to 150 mL. Add HCl until pH=8.0. 0.5 M EDTA i. Get 3.6 g, dissolve to 20 mL ddH2O. To help dissolve EDTA, add NaOH until pH 8. Put solution in magnetic stirrer until dissolved. TE Buffer (50 ml) i. Use 5 mL of 1.0 M Tris (pH 8), add 0.2 mL 0.5 M EDTA (pH 8) then adjust with ddH2O until 50 mL. 50 mL ice-cold TBS i. Recipe: 25 mM Tris pH 8, 0.15 M NaCl f. g. ii. Measure 1.25 mL of 1 M Tris, and 0.44 g NaCl then dilute to 50 mL ddH2O. Extraction buffer i. Use 0.2 mL of 1.0 M Tris pH 8, add 4 ml 0.5 M EDTA (pH 8), add 0.5 g SDS, then adjust with dd H2O until 20 mL. Lysis solution: Recipe is for 0.75 ml. Create 10mL and aliquote in microfuge tubes. i. 6 M Guanidine-HCl 0.43 g of the solid Guanidine-HCl ii. 0.1 M Sodium Acetate Measure 0.0062 g iii. dilute all to 0.75 mL of ddH2O. Sources Extraction of DNA from onion. Access Excellence. Lane, Jo Anne. (Accessed 1 August 2008) Available at: http://www.accessexcellence.org/AE/AEPC/WWC/1994/extraction_of_dna.php. Sambrook, J., E.F. Fritsch, and T. Maniatis, Molecular Cloning: A Laboratory Manual. 2nd ed. Vol. 1 and 2. 1989, United States of America: Cold Spring Harbor Laboratory Press.