Supplementary file1
advertisement

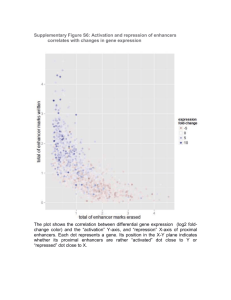
Estimating a useful flanking region size to screen for regulatory elements: mining NCBI databases for distribution of known gene regulatory regions. Recent reports (1,2) of gene-centric computational approaches for identification of putative regulatory regions have limited upstream region search space to 10 kb. Most potential regulatory sequences would be missed using a more traditional approach limited to studying only a few kilobases upstream of the transcription initiation site. An additional consideration is that many genes have poorly identified first exons and that first introns can exceed 10 kb. To develop a more systematic approach to the estimation of generally required flanking base pair regions for computational screening, we mapped known regulatory regions using NCBI Entrez resources. To do this, we used the EntrezNucleotide query (“[FKEY] AND human[ORGN]” and “[FKEY] AND mouse[ORGN]” for human and mouse respectively), we downloaded all the NCBI GenBank sequences (209 human and 136 mouse) that had an explicitly defined regulatory region. Because some of the sequences had more than one enhancer defined, the 209 human sequences had 239 enhancers, while the mouse sequences had 141. The “.gbk” (genbank format) files of each these sequences were then parsed to extract the enhancer regions as defined in the “feature” section of the individual gbk files and then mapped to its corresponding gene and calculated for the relative distance of these enhancers with respect to the associated gene transcriptional start site. In all cases where the enhancer regions described in the gbk files were less than 17 bp, flanking regions of 15 bp (upstream, downstream or both) were added, depending upon the original sequence, to facilitate blat search (for blat search, the minimum length of query sequence should be 17 bp). Thirtytwo of the 239 enhancers were localized to an intron while 192 were mapped to the upstream region of the respective gene. Four of them were mapped to exonic regions of which 3 (enhancers of genes MNDA, OTC and ISG20) of them occur in 5’UTR while one of them maps to a coding exonic region of gene GRIN1. Eleven were found to be occurring downstream to 3’ region. A significant number (76%) were found within upstream 10 kb region. This is of interest in the light of recent findings in Drosophila wherein all the true-positive predictions were within 10 kb of a known or predicted TSS of a gene whose expression was regulated by five TFs involved in anterior-posterior embryonic patterning (3). The mouse enhancers also behaved in similar fashion (Figure 1). Based on these results, we thought, on average, 40 kb flanking region was a reasonable space to search for regulatory signals even though there are instances where regulatory regions are known to occur in regions as far as 100 thousand base pairs upstream or downstream. In some cases, promoters for genes not affected by the cisregulatory module (CRM) are located between the CRM and its target (4). For example, the cis-regulatory region that directs mouse Pax6 expression in the developing pretectum, neural retina and olfactory region, located approximately 77 kb downstream of the 3’ polyA-addition site of the PAX6 gene, actually resides within the transcription unit of an unrelated neighboring gene PAXNEB (5). Therefore, the formidable challenges of “how much” flanking regions to be searched and the association of the predicted regulatory regions with their gene targets still remain and can be met only by experimental validation. Our present results of known human and mouse enhancer mappings relative to TSS and the recent reports of all true-positive Drosophila developmental enhancers occurring within 10 kb of a known or predicted TSS (3) can add additional insights to the predicted putative regulatory regions and their gene associations. Proximity is not always a reliable indicator of the target of a predicted CRM, and a considerable amount of DNA around any gene needs to be examined for a potential role in regulation (4). Thus, our approach of increasing the flanking base pairs’ length and making available precomputed potential regulatory regions for the same provide researchers an “extra space” to work with especially in cases where the immediate flanking regions lack any critical regulatory regions. The problem remains though for cases like PAX6, cited above, in which their regulatory regions are known to occur further up and downstream of the gene and thus lay outside the flanking base pair regions of 40 kb. In these cases the manually curated TraFaC server and similar servers can still be used to identify highly conserved regions after including larger flanking base pair regions in the analysis. REFERENCES 1. 2. 3. 4. 5. Jegga, A.G., Sherwood, S.P., Carman, J.W., Pinski, A.T., Phillips, J.L., Pestian, J.P. and Aronow, B.J. (2002) Detection and visualization of compositionally similar cis-regulatory element clusters in orthologous and coordinately controlled genes. Genome Res, 12, 1408-1417. Dieterich, C., Cusack, B., Wang, H., Rateitschak, K., Krause, A. and Vingron, M. (2002) Annotating regulatory DNA based on man-mouse genomic comparison. Bioinformatics, 18 Suppl 2, S84-90. Berman, B.P., Pfeiffer, B.D., Laverty, T.R., Salzberg, S.L., Rubin, G.M., Eisen, M.B. and Celniker, S.E. (2004) Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol, 5, R61. Miller, W., Makova, K.D., Nekrutenko, A. and Hardison, R.C. (2004) Comparative genomics. Annu Rev Genomics Hum Genet, 5, 15-56. Griffin, C., Kleinjan, D.A., Doe, B. and van Heyningen, V. (2002) New 3' elements control Pax6 expression in the developing pretectum, neural retina and olfactory region. Mech Dev, 112, 89-100. Distance Relative to TSS Figure 1: Distribution of human (red) and mouse (blue) enhancers with respect to the transcription start of the nearest neighboring gene. 100000 90000 80000 70000 60000 50000 40000 30000 20000 10000 0 -10000 -20000 -30000 -40000 -50000 -60000 -70000 -80000 -90000 -100000 No. of Enhancers (human/mouse)