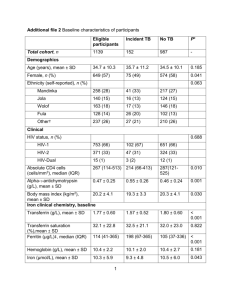
Methods S2
advertisement

Methods S2. Quantification of phospholipids and glycolipids by quadrupole timeof-flight mass spectrometry One lipid aliquot was dissolved in 500 µl of Q-TOF solvent (methanol/chloroform/300 mM ammonium acetate, 665:300:35, v/v/v; Welti et al., 2002). 20 µl of this lipid solution was added to 160 µl Q-TOF solvent and 20 µl of the internal standard mixture. The internal standard mixture contained in 20 µl: 0.2 nmol of each di14:0-PC, di20:0-PC, di14:0-PE, di20:0-PE, di14:0-PG, di20:0-PG, di14:0-PA and di20:0-PA; 0.03 nmol di14:0-PS and di20:0-PS; 0.3 nmol of 34:0-PI; 0.15 nmol 34:0-MGDG, 0.10 nmol 36:0-MGDG; 0.2 nmol 34:0-DGDG, 0.39 nmol 36:0 DGDG; 0.4 nmol SQDG. All phospholipids were obtained from Avanti Polar Lipids. Galactolipids, SQDG and soybean PI were obtained from Larodan and were hydrogenated to remove double bonds (Buseman et al., 2006). Lipids were quantified using a Q-TOF (quadrupole time-of-flight) mass spectrometer (Q-TOF 6530; Agilent Technologies). The samples were injected into the mass spectrometer in chloroform/methanol/300 mM ammonium acetate (300:665:35, v/v/v; Welti et al., 2002) at a flow rate of 1 µl min-1 using a chip-based nanospray ion source (HPLC Chip/MS 1200 with infusion chip; Agilent Technologies). Lipids were ionized in the positive mode with a fragmentor voltage of 270 V. Molecular ions (ammonium or proton adducts; Devaiah et al., 2006) were selected in the quadrupole and fragmented in the collision cell with nitrogen gas using fragmentation energies shown in Table S1. Data were processed with the Mass Hunter Workstation software (Version B.02.00; Agilent Technologies). Quantification was achieved after extraction of neutral loss or precursor ion scanning data from the MS/MS fragment spectra (Devaiah et al., 2006; Table S3). Data were corrected for isotopic overlap, and nmol values were calculated using a linear regression of the two internal standards employed for each lipid class (Welti et al., 2002; Devaiah et al., 2006). References Buseman, C.M., Tamura, P., Sparks, A.A., Baughman, E.J., Maatta, S., Zhao, J., Roth, M.R., Esch, S.W., Shah, J., Williams, T.D. and Welti, R. (2006) Wounding stimulates the accumulation of glycerolipids containing oxophytodienoic acid and dinor-oxophytodienoic acid in Arabidopsis leaves. Plant Physiol. 142, 28–39. Devaiah, S.P., Roth, M.R., Baughman, E., Li, M., Tamura, P., Jeannotte, R., Welti, R. and Wang, X. (2006) Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dα1 knockout mutant. Phytochem. 67, 1907–1924. Welti, R., Li, W.Q., Li, M.Y., Sang, Y.M., Biesiada, H., Zhou, H.E., Rajashekar, C.B., Williams, T.D. and Wang X.M. (2002) Profiling membrane lipids in plant stress responses: role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277, 31994–32002.




![Michaelis Menten Plot Lineweaver-Burke Eadie Hofstee [S] (mM](http://s3.studylib.net/store/data/006783200_1-6f852f20ab04d5dae16be5528b1f30cf-300x300.png)