VIBRATION-ROTATION SPECTRUM OF GASEOUS HCl
advertisement

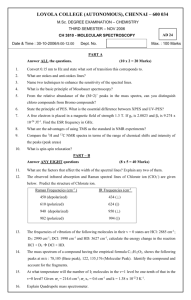
VIBRATION-ROTATION SPECTRUM OF GASEOUS HCl OBJECT: The purpose of this experiment is to introduce you to molecular spectroscopy and to the molecular parameters and thermodynamic properties which can be obtained by analyzing the vibration-rotation spectrum of gaseous hydrogen chloride. REFERENCES (in addition to the Chem 158a,b textbooks): F. Daniels, R. A. Alberty, J. W. Williams, C. D. Cornwell, P. Bender, and J. E. Harriman, Experimental Physical Chemistry, 7th. ed., McGraw-Hill, New York, 1970, pp. 247-256. M. Karplus and R. N. Porter, Atoms and Molecules, Benjamin, Menlo Park, CA, 1971, Ch. 7. EXPERIMENTAL: You will measure fundamental and first overtone vibrational transitions of gaseous hydrogen chloride on a Nicolet Fourier transform infrared (FTIR) spectrometer. The instructor will issue two 10.0 cm cylindrical cells, a blank (empty) and one containing a mixture of HCl and DCl. You will only measure the spectra of the HCl species. When handling the cells, care must be taken to avoid scratching the cell windows. Fingerprints will permanently damage the optical surfaces if they are not washed off immediately with a good quality solvent. The FTIR is controlled by a PC running under Windows 98. The software is all menu-driven and is straightforward to use if one follows the prompts. 1) Preparation of the instrument. The MCT detector on the Nicolet must be cooled with liquid nitrogen. Using the funnel provided, fill the trap for the detector. The fill hole is accessed by opening the small circular door on the left side of the bench. There are two fill holes. Use the one towards the back that is NOT taped shut. Do not overfill as liquid nitrogen spilling over the top of the instrument might cause damage. Check to see that the neutral density filter labeled “screen A” is installed. The detector is very sensitive and the infrared radiation from the source must be reduced in the case of normal, fairly optically transparent samples. Otherwise, the detector is blinded by excess radiation. 2) Start the software by clicking on the Omnic E.S.P. 5.1 icon (not the EZ Omnic icon). If the icon is not on the desktop, go through the following sequence of mouse clicks: Start, Program, Omnic E.S.P. 5.1 folder, Omnic E.S.P. 5.1. Once the program starts, a window will appear that will allow you to select the methods file for the experiment. Select the method "HCl” and click OK. You will probably not need to change the parameters stored in this file. For the record, they are as follows: number of scans, 64; resolution, 0.5 cm-1; format, Absorbance; correction, none; gain, Autogain; velocity, 1.8988; detector, MCT/A; beam splitter, KBr; range, 7500-1700; zero-filling, none; apodization, boxcar; phase correction, Mertz. 3) Initialize the bench for the method that you selected (i.e. HCl) by the clicking on “Collect” and then “Experiment Setup”. The Collect menu that displays the values of the acquisition and processing parameters will appear. The Diagnostic tab accesses useful diagnostic functions that the instructor will demonstrate. You have the option now of changing parameters. If you do so, please use a method name other than HCl. After you have viewed the values of the parameters, click on the OK button to configure the bench. 4) Collect a background spectrum by loading the empty blank cell and clicking on the “Col Bkg” button. While the instrument accumulates the data from the 64 acquisitions, the PC calculates the spectrum from the accumulated interferogram on the fly. The sharp bands are due to atmospheric carbon dioxide and water. Note that the cell heavily absorbs in certain regions of HCl - 1 VIBRATION-ROTATION SPECTRUM OF GASEOUS HCl the spectrum as it is fabricated from a form of glass and hence is infra-red absorbing. These absorbing features in the spectrum will be removed by the computer via background subtraction. This approach would not be possible on a conventional CW spectrophotometer. Normally the cell windows must not be infra-red absorbing. 5) After the background spectrum has been measured, replace the blank cell with the sample cell and click on the “Col Smp” button. When the interferogram of the sample spectrum has been acquired, the difference spectrum will be calculated and displayed on the screen. We recommend that you save the results at this point by clicking on File and Save As. 6) The final step is the measurement of the transition frequencies and the assignment of quantum numbers to each peak. We recommend starting with the bands of the fundamental spectrum of HCl. They are the strongest features in the spectrum and are located at ca. 3000 cm-1. Recall that for each vibrational transition, the R branch for which J increases by one lies to higher frequency than the P branch for which J decreases by one. Also, the transitions for HCl-35 and DCl-35 are more intense by a factor of 3:1 than those of HCl-37 and DCl-37, respectively, and also lie to slightly higher frequency. There are two options for expansion. One can draw a box around the region of interest by holding down the left mouse button and dragging the mouse. Once the box enclosing the region of interest has been drawn, move the cursor inside the box and perform one left mouse click to expand the box. Alternatively, one can use the utilities of the View Finder at the bottom of the Omnic window to expand the frequency scale and then clicking on “View” and “Full Scale” in order to expand along the absorbance (y axis) to fill the screen. To display the full spectrum after expanding it, double click in the white (displayed) portion of the View Finder window. Once a region has been expanded, you can automatically measure the frequencies of the peaks in the window with the peak finder function. Click on “Analyze” and “Find Peaks” in the Analyze submenu. You may need to lower the threshold to obtain the frequencies of weaker peaks or raise the threshold to suppress annotation of noise. The threshold in indicated by a horizontal black line across the spectral window. To raise the threshold, click just above the black line; to lower it, click below. Alternatively, you can manually determine the frequencies with the Annotation Tool symbolized by the large capital T at the bottom of the screen. To enter annotation mode, click on the T. To annotate, move the cursor now labeled by a T so that the arrow is above the peak. Hold down the shift key and perform a left mouse click. The software will find the frequency of the peak closest to the position of the peak. If you simply left-mouse click, the annotation tool will print the frequency of the exact location of the arrow when you click. Once you have annotated a section of the spectrum, you can create and print a report by clicking on “Report” and then on “Preview/Print Report” in the Report submenu. Once the report appears, click on the Print button to print the report. Alternatively, you can start up Microsoft Word that is installed on the PC and use the Edit and Copy functions in Omnic to transfer an image of the screen to the Windows Clipboard. Then enter Microsoft Word and use the Edit and Paste function to transfer the contents of the Clipboard to the Word window that can be saved as a .doc file. We recommend that you insert a page break after each screen dump so that the image of each screen appears on a separate page. 7) Once you have analyzed the components of the fundamental spectrum for the two isotopic species of HCl, turn your attention to the fundamental spectrum of DCl. You should be able to HCl - 2 VIBRATION-ROTATION SPECTRUM OF GASEOUS HCl predict its approximate location by using the well known dependence of the vibrational frequency on reduced mass. Unfortunately, you will only obtain the transitions for the R branch as the cell window is totally opaque in the region of the P branch. 8) Next turn your attention to the analysis of the first overtone spectrum of DCl whose position can be predicted by the harmonic oscillator approximation. 9) Finally analyze the first overtone spectrum of HCl whose approximate location is also predictable from the harmonic oscillator approximation. These bands partially overlap broad, but fortunately weak, artifacts that should not be confused with authentic transitions. CALCULATIONS: As shown in Daniels et al. and in Karplus & Porter, the vibrational-rotational energy of a diatomic molecule is given by equation (1): E = (v + 0.5) - x(v + 0.5)2 + BJ(J + 1) - DJ2(J + 1)2 - (v + 0.5)J(J + 1) (1) where the subscript e has been dropped from all of the parameters to simplify the format. Since E and all spectroscopic parameters are given in wavenumbers, each energy difference, E, of each allowed transition is given directly by a spectroscopic frequency in wavenumbers. For each spectroscopic transition v"v', J"J' (" denotes the ground state where v" = 0; ', the excited state), the following constraints on v' and J' apply: v' = v" + 1 for the fundamental transition and v" + 2 for the first overtone, J' = J" - 1 for a P band transition and J" + 1 for an R band transition. The energy difference for each allowed transition is given by equation (2): E(v',J',v",J") = E(v',J') - E(v",J") = (v' - v") + x[(v" - v')(v' + v" + 1)] + B[J'(J' + 1) - J"(J" + 1)] + D[J"2(J" + 1)2- J'2(J' + 1)2] + [(v" + 0.5)J"(J" + 1) - (v' + 0.5)J'(J' + 1)] (2). This is a complicated expression but it has some features which permit a direct calculation of all the parameters for each isotopic species. Note that the expression for E depends linearly on each of the parameters. The equation is simply a variation of the well known expression y = mx + b with a few new twists: 1) the intercept is zero, 2) there are 5 rather than 1 independent variables, and 3) each independent variable is a non-trivial, but well defined combination of the quantum numbers v', J', v", and J". For example, the independent variable associated with the parameter B is [J'(J' + 1) - J"(J" + 1)]. Therefore, the dependence of E on [J'(J' + 1) - J"(J" + 1)], holding the other variables constant, yields the parameter B as the slope. In other words, the extraction of the required parameters from the spectroscopic data is simply a logical extension of the method of least squares to the case of several independent variables. The extension is referred to as multivariate analysis. The analysis of the spectral data consists of the following steps: HCl - 3 VIBRATION-ROTATION SPECTRUM OF GASEOUS HCl a) The data from the FTIR are in wavenumber and no correction of the raw data is required. The moving mirror in the FTIR’s interferometer is tracked by a laser that provides an internal calibration for the frequency data. b) Separate the observed peaks into four sets, those for HCl-35, HCl-37, DCl-35, and DCl-37. You will analyze each set separately. c) Assign quantum numbers to the transitions for each isotopic species. d) Calculate the spectroscopic parameters - , x, B, D, and - for each isotopic species via a multivariate analysis (vide infra) of the assigned transitions. Also calculate the bond length, re, from B, the force constant, k, from , and an estimate of the bond dissociation energy from and x. e) Using the molecular constants obtained, the published isotopic abundances (cf. CRC Handbook), and standard statistical mechanical formulae, calculate the following thermodynamic quantities for naturally occurring gaseous HCl at 25.0C: Cp and S. ANALYSIS OF THE DATA USING NCSS: This section discusses how to analyze the data using the NCSS software. This document assumes familiarity with the handout on the use of NCSS and general spreadsheet techniques. The first step is to prepare a spreadsheet for each isotopic species that contains the data and the quantum numbers of the transitions. Each row of the spreadsheet contains information on a single transition. Consider organizing the spreadsheet in blocks with sections for the P branch of the fundamental, the R branch of the fundamental, the P branch of the overtone, and the R branch of the overtone. The first column should contain the transition energies in wavenumbers. The next four columns contain the quantum numbers in the order v", v', J", and J'. The task of entering the quantum numbers can be simplified if one makes liberal use of copying and pasting. Use all your data. For example, if you have the R branch transition and not the P branch, still use the R branch datum. Once the data for the regression have been entered, save the spreadsheet. In the next step, calculate using spreadsheet techniques the independent variables for the five parameters to be determined from the spectroscopic data: item 1 2 3 4 5 parameter e exe Be De e corresponding independent variable (v'-v") (v"-v')(v'+v"+1) J'(J'+1)-J"(J"+1) J"2(J"+1)2-J'2(J'+1)2 (v"+0.5)J"(J"+1)-(v'+0.5)J'(J'+1) Consider checking a few entries with a result calculated manually so that you know that the variables have been defined correctly. Once the full spreadsheet has been defined, re-save it and prepare for the regression by clicking on the Regression/Correlation item in the Analysis menu and selecting the Multiple Regression module. The handout on the use of the regression modules in NCSS discusses how to use the procedure. The following special features apply to this experiment. There is no intercept so the Remove Intercept option should be set to Yes. In defining the independent variables, select all five which are located in columns 6-10. The Correlations report is useful as it contains the full correlation matrix from which the full covariance HCl - 4 VIBRATION-ROTATION SPECTRUM OF GASEOUS HCl matrix can be calculated. Also pay very close attention to the table of residuals for each regression calculation. The accuracy and precision of the FTIR is ca. 0.1 cm-1. Errors in excess of this indicate some type of error, e.g. input error, incorrect assignment, mistake in setting up the spreadsheet. The spectroscopic parameters are all positive. With the exception of exe and the centrifugal distortion constant, the relative errors should be small. Large relative errors are indicative of a calculational or measurement error. The table of residuals is often helpful in locating the error. REPORT: Include the following with your short report: the report sheets, all spectra with assigned transitions, the NCSS spreadsheets and regression reports, notebook entries, all relevant printouts, and sample calculations. Report all spectroscopic parameters and their associated 95% confidence intervals to the correct number of significant digits. Do not calculate the random error of the bond dissociation energy. Systematic errors in the calculation are much larger than the random errors that your analysis would address. Similarly do not calculate uncertainties for S and Cp. However, use a propagation of errors analysis to calculate an uncertainty for r e and k for each isotopomer. LONG REPORT. If you select this experiment for a long report, write it in the style of a short paper in the Journal of Physical Chemistry. The report should contain the following elements: a) A brief introduction. What have you done and what will you discuss? b) Experimental. What sample was used? which instruments? Do not go into detailed procedures, but provide instrumental details that one would need to repeat the experiment. c) Results. Provide a table of the assigned frequencies and give a brief (few sentences) rationale for the assignment. An appropriately labeled figure of the spectrum (cut and paste and label) with a well chosen caption might make the point better than extensive text. Briefly describe how the spectroscopic parameters were obtained. Briefly comment on the errors in the results and the internal consistency of the results (i.e. can you estimate one parameter from the others). Don't go into great detail on the theory of quantum mechanics and molecular spectroscopy. You are writing a paper on HCl, not a monograph on physical chemistry. d) Discussion. This is the most important section of the paper. Discuss what you have learned about the molecule HCl. How stable is the molecule? Your data provide tests for the harmonic oscillator, rigid rotor, and Born-Oppenheimer approximations (all 3) and this is worthy of discussion. e) Literature. Compare your spectroscopic parameters with comparable results from the chemical literature. Use of the JANAF Thermochemical Tables and Huber and Herzberg’s compilation will ease your work. f) A brief conclusion. With this, you have all the elements of effective communication: tell them what you're going to tell them, tell them, and tell them what you told them. hcl_2001.doc, FJG, revised 3 Jan. 2002 HCl - 5 VIBRATION-ROTATION SPECTRUM OF GASEOUS HCl REPORT SHEET Date expt. completed: _____________ Date report due: _____________ NAME:_________________________________ Date report submitted: _____________ I) Spectroscopic parameters for the isotopomer H35Cl Experimental Literature Values Be (cm-1) ___________________ ___________________ e (cm-1) ___________________ ___________________ De (cm-1) (cent. dist. constant) ___________________ ___________________ e (cm-1) ___________________ ___________________ exe (cm-1) ___________________ ___________________ De = e2/4exe (cm-1) (bond dissn.energy) ___________________ ___________________ re (Å) ___________________ ___________________ k(N/m) ___________________ II) Spectroscopic parameters for the isotopomer H37Cl Experimental Be (cm-1) ___________________ e (cm-1) ___________________ De (cm-1) (cent. dist. constant) ___________________ e (cm-1) ___________________ exe (cm-1) ___________________ De = e2/4exe (cm-1) (bond dissn.energy) ___________________ re (Å) ___________________ k(N/m) ___________________ N.B. ADDITIONAL INFORMATION IS REQUESTED ON THE BACK OF THIS FORM! HCl - 6 VIBRATION-ROTATION SPECTRUM OF GASEOUS HCl III) Spectroscopic parameters for the isotopomer D35Cl Experimental Literature Values Be (cm-1) ___________________ ___________________ e (cm-1) ___________________ ___________________ De (cm-1) (cent. dist. constant) ___________________ ___________________ e (cm-1) ___________________ ___________________ exe (cm-1) ___________________ ___________________ De = e2/4exe (cm-1) (bond dissn.energy) ___________________ ___________________ re (Å) ___________________ ___________________ k(N/m) ___________________ IV) Spectroscopic parameters for the isotopomer D37Cl Experimental Be (cm-1) ___________________ e (cm-1) ___________________ De (cm-1) (cent. dist. constant) ___________________ e (cm-1) ___________________ exe (cm-1) ___________________ De = e2/4exe (cm-1) (bond dissn.energy) ___________________ re (Å) ___________________ k(N/m) ___________________ V) Molar thermodynamic parameters (naturally occurring mixture of HCl35 and HCl37) at 25C. Literature Values S (J/K-mole) ________________________ ___________________________ Cp (J/K-mole) _______________________ ___________________________ HCl - 7