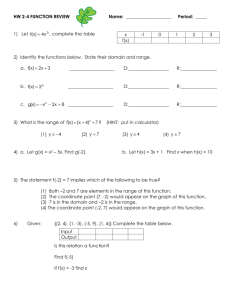
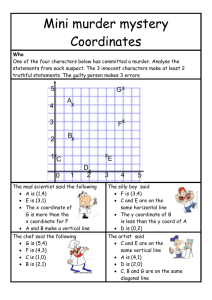
Supplementary Information (doc 5447K)
advertisement

Supplementary Information Material and methods 5 Study floodplains The Val Roseg catchment is situated in the eastern Swiss Alps and belongs to the lower austroalpine Bernina nappe (9°53’53’’E, 46°29’24’’N). Geology consists of crystalline bedrock composed mainly of diorite and granite (Malard et al., 1999). It covers an area of 66.5 km2 from which 30.1% is glaciated. Annual precipitation rates are approximately 1.6 m, with around 50% 10 as snow. The altitude of the catchment ranges from 1766 to 4049 m a.s.l., and the study area ranged from 2009 to 2336 m a.s.l. The Roseg River is an 11.3-km long second-order tributary of the river Inn, which drains into Danube. Average annual discharge and water temperature are 28.5 m3 s-1 and 3.6 °C (range0.1 to 12.6°C). Approximately 30% of the water volume of the Roseg River is fed by water 15 from two valley glaciers, the Roseg glacier and Tschierva glacier, both of which have retreated continuously over the last century (Maisch 1988, Tockner et al., 1997, Malard et al., 2000, Tockner et al., 2002, BAFU 2010). Permanent flowing first-order tributaries contribute groundwater and snowmelt to the Roseg with peak flows during June and August (Malard et al., 2000). The channel network within the floodplain shows a distinct contraction in October caused 20 by the freezing of glacial water. The Loetschental catchment is in the Rhone-Valley in the southwest part of the Swiss Alps (07°49'03''E, 46°25'08''N), harboring the second-order kryal stream Lonza which drains into the Rhone. It is part of the old crystalline Aare massif that is dominated by amphibolite and gneiss (Labhart 1998). The altitude of the 77.8 km2 valley ranges from 1375 to 3200 m a.s.l. 25 (Schmidt et al., 2009, BAFU 2010), and the study area was located between 1929 to 2210 m a.s.l. Approximately 36.5% of the valley is glaciated by the Lang glacier and the Jegi glacier. The 1 River Lonza shows an average discharge and water temperature of 37.2 m3 s-1 and 4.0 °C (range of 0.1 to 10.9 °C)(BAFU 2010). The kryal tributary fed by the Jegi glacier (Anunbach) and several first-order krenal tributaries drain into the Lonza within the study area. Most tributaries 30 run dry during October and the channel network experiences a similar periodical contraction as the Roseg cannel network. The Macun Lakes region is a high alpine cirque situated in the Swiss National Park, and located in the mid-eastern part of the Swiss Alps (10°07'31''E, 46°43'51''N). It belongs to the upper austroalpine Silvretta nappe and geology consists mainly of crystalline rock dominated by 35 orthogneiss. The catchment is divided into a southern and northern basin that differs in their water source (Robinson and Matthaei 2007). The northern basin is mainly groundwater and snowmelt fed, whereas the southern basin is fed mostly by rock glaciers. This catchment also experiences contraction of surface channels in October (Robinson and Matthaei 2007). 40 Details on bacterial total cell numbers A 0.5 ml aliquot of collected sediment (n=3) was suspended in 1.11 ml paraformaldehyde (2%, final concentration) in an Eppendorf tube and fixed for 24 h at 4°C followed by three washing steps with 1 x PBS and 5 min centrifugation at 10,000 g between washing steps. Samples were then stored at -20°C in a 1:1 mix of PBS/ethanol until further processing 45 (Pernthaler et al., 2001). Weight of the sediment, tube and the storage solution was measured. Attached bacteria were brought into suspension by sonication (Branson Digital Sonifier 250, Danbury, USA, 5-mm tapered microtip, actual output of 20 W) using 1-s sonication pulses for 30 s. After vortexing the sample for 7 s, a short spin centrifugation using a table top centrifuge for 5 s was performed to remove coarse particles interfering with downstream sample processing. 50 The supernatant was transferred into a new Eppendorf tube and served as a template for total cell counting of 4’,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich Co) stained cells. The remaining sediment was dried for 24 h at 60°C and weighed with and without the Eppendorf. 2 A 10 to 60 µl aliquot of template solution was pipetted into 5 ml sterile ultrapure water (MQ) and stained with DAPI (1 µg ml-1 final concentration) for 7 min followed by filtration onto a 55 black polycarbonate filter (0.2-µm pore size, 25-mm diameter, Millipore, Molsheim, GTBP02500) by applying a gentle vacuum (Porter and Feig 1980). Filters were air-dried and embedded into citifluor AF1 (Linaris Biologische Produkte, Wertheim,Bettingen). An epifluorescence microscope (Leica Micorsystem, DMI6000b) was used to take a minimum of 16 photographs of each stained filter to ensure an equal distribution and an appropriate number of bacterial cells (800+) for 60 counting. Photographs were analyzed using the CellC software (Selinummi et al., 2005) or counted manually if background fluorescence interfered with the automated counting routine. Cell numbers were then standardized to the dry mass of the initially suspended sediment by using the weights determined during procession. 65 Details on enzyme assays Approximately 10 g of sediment sample (n=3 per sample) were dissolved in 10 ml MQ and vortexed for 1 min to suspend the associated biofilms. A 150-µl aliquot was then transferred into a 96 well microplate and 100 µl of a 1 mM substrate stock solution was added to get a final substrate concentration of 400 µM (Findlay et al., 2001). The remaining sediment and MQ were 70 dried at 60°C for 48 h to measure the dry weight subsequently used to calculating the OM content. Fluorimetric enzyme assays were performed directly after adding the substrate for up to 24 h using a microplate reader (Tecan Infinite® 200, Switzerland). The excitation wavelength was set at 365 nm and fluorescence emission was measured at 445 nm. Plates were incubated on a plate shaker at 15°C between measurements. All fluorescence values were corrected for 75 quenching by adding known quantities of free Methylumbelliferone (MUF) to the samples and to MQ and bicarbonate buffer blanks. Reaction rates were calculated using the linear part of the fluorescence reaction curve. Potential enzyme activities were standardized to nmol substrate g-1 OM h-1. 3 Details on bacterial community fingerprinting 80 Bacterial community structure was assessed by ARISA. Frozen samples (n=2 per sample) were extracted using the PowerSoil DNA isolation Kit (MoBio, Carlsbad) following manufacturer’s instructions. DNA was amplified using the fluorescein (6-FAM) labeled universal forward primer 1406f-6FAM (16S rRNA gene, 5’-FAM-TGYACACACCGCCCGT-3’, Y=T,C) and the 85 bacteria specific reverse primer 23Sr (5’-GGGTTBCCCCATTCRG-3’, B=G,T,C, R=G,A)(Yannarell et al., 2003). PCR was performed using a TProfessionalthermocycler (Biometra GmbH, Göttingen) in a final reaction volume of 25 µl with a mix of 1x GoTaq®Flexi buffer, 3 mM MgCl2, 0.25 mM each of dNTP, 0.05 U µl-1 of GoTaq®Flexi DNA Polymerase (Promega, Switzerland), 0.25 mg ml-1 bovine serum albumin (Sigma-Aldrich, Switzerland), 0.4 µM of each primer (Microsynth, Switzerland), and 1 µl of template DNA. The reaction mix was 90 initially denatured for 2 min at 94°C followed by primer annealing at 55°C for 35 s and extension of 2 min at 72°C. Denaturation time was then reduced to 35 s and the cycle was repeated 29 times followed by a final extension for 2 min at 72°C. ARISA fragment analysis was performed as described in Bürgmann et al. (2011). Briefly, a 1 µl aliquot of PCR product was mixed with 9 µl of HiDi formamide and 0.5 µl Liz1200 size standard (Applied Biosystems, Switzerland) and 95 denatured prior to capillary electrophoresis on a PCR thermocycler for 3 min at 95° C and subsequently placed on ice. Denaturing capillary electrophoresis was performed on a 3130XL Capillary Genetic Analyzer (Applied Biosystems, Switzerland) equipped with a 50-cm capillary using POP-7 polymer. ARISA fragments were analyzed with the Southern size-calling method with a background cut-off level of 50 fluorescence units. Binning of peak areas was done with 100 automatic and interactive binning R scripts (Ramette 2009) leading to relative fluorescence intensities (peak areas) between 197 and 1400 bp. The mean of the relative fluorescence intensities of the extracted samples (n=2) was used for subsequent analysis. These fluorescence intensities were Wisconsin double standardized prior to analysis (relative peak 4 area of an operational taxonomic unit (OTU) of a sample is first standardized by maxima peak 105 area of this OTU within all samples and then by site total OTUs relative fluorescence intensity). Details on data analysis Environmental factors, cell abundance was tested for homogeneity using a Bartlett-Test. Comparison of environmental variables, cell abundance and enzymatic activity between 110 catchments, water source and sampling date were done using three-factor ANOVA (Type I SS) followed by Tukey’s HSD when differences were significant. Catchment, water source and sampling date were treated as fixed factors in the ANOVA. Akaike information criterion (AIC) model reductions were performed for confirmation of non-significant model terms. Normality of residuals was assured by performing a Shapiro Willk’s test and examining the QQ-plot of the 115 residuals. If one of the assumptions was violated, data were transformed, after checking with the Box-Cox-Method (Krambeck 1995), by ln x 1 or x Max log Likelihood . Percentage values were arcsin x transformed prior to analysis. In case of non-conformity after transformation, a non- parametric Kruskal-Wallis test followed by multiple comparisons was performed instead of ANOVA. Tukey like Post-hoc tests were performed for boxplots independent of conformity based 120 on linear models compared with the glht() function available in the mulitcomp package in R. P-values were holmes adjusted. Community fingerprinting results, enzymatic activities and environmental data were analyzed using NMDS in combination with vector and factor fitting. Untransformed enzyme activities and environmental parameters were used. Significance of fitted vectors and factors 125 was tested by a permutation test (999 permutations) and visualized as biplots and dispersion ellipses in the NMDS ordinations (Figures S4 and S5). Generalized additive models were fitted on the NMDS for raw physico-chemical data to check linearity of fitted vectors (biplots) and used to assess the importance of single physico-chemical variables potentially constraining the BCC or EF pattern (Appendix Tables 6 and 7) (Bennion et al., 2011). Physico-chemical variables were 5 130 therefore interpolated with the function ordisurf() from the vegan package in R (Oksanen et al., 2011). Shortly, the function fits a smooth surface using thinplate splines and the percentage of variance of each factor explained by the surface can be seen as a measure of how well an environmental variable explains the a priori and unconstraint NMDS pattern. The advantage of this approach is that linear projections of environmental variables (biplots) in the ordination 135 space may not be de facto ideally linear as i.e. in constraint ordinations, thus takes non-linear relationships better into account. The totally explained variance of the physico-chemical factors on BCC and EF was assessed by a global RDA model incorporating all samples and physico-chemical parameters. Canonical coefficients (i.e. the regression coefficients) of the first two axes and R2 of forward 140 selected explanatory variables were calculated. This allows assessing which variables are most important to explain EF or BCC structure on these two axes and can be qualitatively compared to the explained deviance of the respective GAM model (Appendix Tables 6 and 7). RDA based variation partitioning were performed to evaluate the influence of assessed chemical and physical (temperature and D90D10) factors on bacteria structure and function as a whole and 145 within the specific water systems. For both approaches, variation inflation factors were calculated and physico-chemical factors which had a value above 20 were removed before a forward selection was performed. This minimizes collinearity of respective variables with other variables included in the analysis. Unique fractions of RDA were tested by an ANOVA like permutation test. Reported are the adjusted R2 (Peres-Neto et al., 2006, Blanchet et al., 2008). 150 RDA based analysis were performed with Hellinger transformed ARISA and enzymatic activity data. Permutational multivariate analysis of variance (PERMANOVA) was used to assess the influence of water source, catchment and sampling date (full factorial model) on community and enzyme activity structure (Anderson 2001). Additionally, a factor fitting for water source, 155 catchment and sampling date ordinations was performed for the NMDS ordinations and tested 6 with a permutation test (999 permutations) leading to the r2correlation coefficient (Appendix Tables 6 and 7). Correlation of single enzymes to physico-chemical factors were tested by multiple linear regression. The models were selected by AIC, significance of single predictors was tested by 160 permutational ANOVA (Type III SS) and relative importance of predictors in the linear model was assessed using the lmg metric (Chevan and Sutherland 1991, Grömping 2006). The enzymatic activities standardized to OM were used for the models without OM as a predictor incorporated. This prevents autocorrelation. Physico-chemical factors were ln x 1 transformed. A pairwise comparison between a specific water source in a specific catchment between 165 specific sampling dates was also performed for EF and BCC by the means of PERMANOVA. These results were then used as a measure for the coupling of BCC and EF. This was done by giving pairwise PERMANOVAS with P-values lower then 0.05 (i.e. significant differences due to a temporal shift in EF or BCC) a value of 0 and PERMANOVAS with P-values higher then 0.05 (i.e. no differences in EF or BCC between two sampling dates) a value 1, respectively, and then 170 summing the corresponding pairs of structure and function. Sums of zero and 2 indicate a potential coupling of structural and functional measures, whereas a value of 1 indicates a decoupled relationship. I.e. if BCC shifts (=0) and EF stays stable between two sampling dates (=1) we would detect this as an uncoupling of BBC and EF (sum=1) (Appendix Table 4). This assessment was supported by a Mantel test and a procrustes analysis. The Mantel test between 175 ARISA and enzyme activities was based on 999 permutations (Mantel 1967). The procrustes analysis of the corresponding NMDS ordinations leads to m2 statistic, which is used as a measure of congruence of the two ordinations, and the procrustes correlation r 1 m 2 . Configurations of NMDS were scaled to equal dispersion. Non-randomness between the two configurations was tested with the function protest by 999 permutations (Gower 1975, Digby and 180 Kempton 1987, Jackson 1995, Peres-Neto and Jackson 2001). Procrustes analysis was 7 additionally performed for ARISA and enzymatic activities vs. environmental variables NMDS ordinations (Appendix Table 8). Multivariate homogeneity of groups dispersion (MHGD), as an analogue of Levene’s test for homogeneity of variance, was performed to assess changes in beta diversity (i.e. changes of 185 the changes in community structure; 2nd order variation) of fingerprinting profiles and the changes in variation of enzyme activities, and physico-chemical characteristics. Generally, multivariate dispersion (variance) can be calculated by the distances of group members to a group centroid in multivariate space. Differences in dispersions can then be tested by ANOVAs. Bray-Curtis dissimilarities of transformed and standardized datasets were reduced from their 190 original distances to principal coordinates (PcoA) which embeds them within an Euclidian space. The average distances to centroids were then calculated within groups, thus are based on principal coordinate axes rather than the original distances. Differences between groups were assessed using a permutational ANOVA test (999 permutations) followed by a Tukey’s HSD (Figures S3, S9) (Anderson et al., 2006). NMDS, MHGD and PERMANOVA were all based on 195 Bray-Curtis dissimilarity matrices calculated from the Wisconsin double standardized relative fluorescence intensity of ARISA profiles, Wisconsin standardized square-rooted enzyme activities and ln(x+1) transformed environmental variables (Bray and Curtis 1957). Shannon diversity index was calculated for OTU’s (operational taxonomic units). All analyses were done using the vegan, relaimpo, stats and mgcv package in R (R 200 Development Core Team, 2011, Oksanen et al., 2011). Results Detailed results of physico-chemical parameters 205 Additional physico-chemical variables are depicted in Figure S2.There was a strong influence of catchment, water source and sampling date on water temperature (ANOVA: 8 F2,89=18.53, F1,89=32.98 and F2,89=8.40, respectively, P<0.001). Water temperatures in the three catchments showed similar patterns. Kryal (glacial) systems were colder (range: 0.3 to 11.9°C, mean: 4.3°C) during the study period than groundwater (krenal) systems (range: 2.3 to 20.7°C, 210 mean: 8.5°C, Tukey’s HSD: P<0.001). Val Roseg streams were generally colder than streams in the other two catchments (Tukey’s HSD: P<0.001). Temperature was lower in October compared to August and June (Tukey’s HSD: P<0.05). The krenal system in Val Roseg had the lowest mean temperatures compared to Macun and Loetschental krenal sites, which did not differ (5.8 vs. 10.1 and 10.5°C, respectively, ANOVA: F2,52=5.91, P<0.001, Tukey’s HSD: 215 P<0.05). Conductivity showed significant interactions between catchment and water source, catchment and sampling date and water source and sampling date (ANOVA: F2,89=5.70 and F3,89=2.90 and F2,89=5.80 respectively, P<0.05). There were significant differences in conductivity between kryal and krenal systems in the Loetschental (mean krenal: 89.9±48.5, mean glacial 220 water: 44.74±23.3 µS cm-1, Tukey’s HSD: P<0.01). Lowest conductivity values were found in the Macun catchment (mean: 7.9 ± 3.4µS cm-1, Tukey’s HSD: P<0.01) and highest values in Loetschental and Val Roseg, which did not differ (69.8 ± 44.9 and 62.6 ± 34.5 µS cm -1, respectively, ANOVA: F2,89=200.76, P<0.001, Tukey’s HSD: P=0.64). The three catchments were clearly distinct in respect to sediment pH with lowest mean 225 pH measured in Macun followed by Loetschental and Val Roseg (mean: pH 4.86, 6.64 and 7.27, respectively, ANOVA: F2,89=558.08, P<0.001, Tukey’s HSD: P<0.001). Sediment pH showed a significant interaction between catchments and water source with a significant difference between water systems (kryal vs. krenal) only in Val Roseg (ANOVA: F2,89=6.82, P<0.01, Tukey’s HSD: P<0.001). 230 Macun had higher amounts of OM (mean: 2.54 ± 2.43% OM dw-1) in the sediment compared to the other two catchments, which were similar (Loetschental: 0.53 ± 0.36% OM dw-1, Val Roseg: 0.55 ± 0.64% OM dw-1, Kruskal-Wallis: H=46.7, df=2, P<0.001, multiple comparison: 9 P<0.01). OM was generally lower in the kryal than in the krenal system (mean: 0.76 and 1.32% OM dw-1, respectively, Kruskal-Wallis: H=19.07, df=1, P<0.01, multiple comparison: P<0.01). 235 DOC was lower in Val Roseg compared to Macun but not to Loetschental, whereas Macun and Loetschental did not differ from each other (mean: 0.95 ± 1.55 mg DOC ml -1, 2.16 ± 3.51 and 2.37 ± 3.93 mg DOC ml-1, respectively, Kruskal-Wallis: H=10.59, df=2, P<0.01, multiple comparison: P<0.01). There was no significant difference in DOC between the two water types in each catchment (Kruskal-Wallis: H=3.33, df=1, P=0.07). October DOC concentrations were 240 higher compared to those in August (Kruskal-Wallis: H=24.69, df=2, P<0.001, multiple comparison: P<0.01). Loetschental kryal DOC concentrations were higher in June than the ones in August (Kruskal-Wallis: H=61.43, df=15, P<0.001, multiple comparison: P<0.01) POC concentrations in Val Roseg differed from Macun but not from Loetschental, (Kruskal-Wallis: H=12.22, dF=2, P<0.01, multiple comparison: P<0.01). More detailed, there was 245 a significant difference in POC in krenal sites between Macun and Val Roseg (mean: 1.22 ±1.34 and 0.21 ± 0.12 mg C l-1, respectively, Kruskal-Wallis: H=21.96, df=5, P<0.001, multiple comparison: P<0.01). There was a significant interaction between catchment and sampling date in TIC concentrations (ANOVA: F3,89=10.26, P<0.001). The Val Roseg catchment showed temporal 250 differences with lower TIC concentrations in August than in October and June (Tukey’s HSD: P<0.01). TIC differed between catchments and was lowest in Macun (mean: 1.77 ± 1.2 mg C l-1) and highest in Roseg and Loetschental (mean: 5.39 ± 2.33 mg C l-1 and 4.02 ± 1.43 mg C l-1, respectively, ANOVA: F2,89=67.90, P<0.001, Tukey’s HSD: P<0.05). Krenal systems had higher TIC values than kryal systems (ANOVA: F1,89=8.12, P<0.01). 255 NH4-N showed no differences between catchments (Kruskal-Wallis: H=0.48, df=2, P=0.79), but lower values for krenal than kryal channels in Val Roseg (Kruskal-Wallis: H=22.63, df=1, P<0.001). 10 NO2-Ndiffered between the two water sources in Val Roseg (krenal mean: 0.53 ± 0.12 µg N l-1, kryal mean: 2 ± 1.04 µg N l-1, Kruskal-Wallis: H=52.66, df=5, P<0.001, multiple comparison: 260 P<0.01). NO3-N did not significantly differ between sites, sampling date and catchments. Macun had highest DON concentrations in the kryal system in October (Kruskal-Wallis: H=41.57, df=15, P<0.001). Macun had higher PN concentrations compared to the other catchments (Kruskal-Wallis: 265 H=19.01, df=2, P<0.001, multiple comparison: P<0.01) and also differences between the two water systems (Kruskal-Wallis: H=9.1, df=1, P<0.01) with highest values in krenal streams. Water systems in Val Roseg also differed in PN concentrations (Kruskal-Wallis: H=10.54, df=1, P<0.01), except that PN was higher in kryal channels. No difference was detected in DP between catchments (Kruskal-Wallis: H=1.78, dF=2, 270 P=0.41). Glacial waters showed highest concentrations of DP (Kruskal-Wallis: H=10.69 ,dF=1, P<0.01), except for Macun where DP was the same for both systems (Kruskal-Wallis: H=0.45, df=1, P=0.50). PP was lower in Macun kryal waters than in the other two catchments (Kruskal-Wallis: H=14.29, df=2, P<0.001, multiple comparison: P<0.01). Kryal systems had higher PP 275 concentrations compared to krenal systems in the Roseg catchment (Kruskal-Wallis: H=22.05, df=1, P=<0.001). Krenal sites had higher PP in Loetschental than in Roseg (Kruskal-Wallis: H=12.93, df=2, p<0.01, multiple comparison: P<0.01). A peak in PP in August was visible in Val Roseg (Kruskal-Wallis: H=8, df=2, p<0.05, multiple comparison <0.05). No difference in PO4-P concentrations was detected between catchments (Kruskal- 280 Wallis: H=2.63, df=2, P=0.27), although glacial waters generally had highest PO 4–P levels in Val Roseg (Kruskal-Wallis: H= 13.88, df=1, P<0.001). The D90/D10 sorting coefficient did not show a significant difference between the catchments, water source or the sampling date respectively (ANOVA: P>0.05). 11 The MHGD analysis showed difference in variation of physico-chemical characteristics 285 between groupings of same water source, sampling date and catchment (Permutation test: F15,89=4.16, P<0.001, Figure S3). There was an interaction of catchment, water source and sampling date on distances to centroids (ANOVA: F3,89=5.71, P<0.01) as well as an interactive effect of catchment and sampling date (ANOVA: F3,89=5.32, P<0.01). Catchment, water source and sampling date had also an effect on the distance to centroids (ANOVA: F2,89=7.16, P<0.01, 290 F1,89=10.98, P<0.01, F2,89=3.77, P<0.01, respectively). In particular, Roseg kryal waters showed higher variation in October compared to most krenal sites, except for Loetschental June and October and Macun August (Tukey’s HSD: P<0.05). Roseg kryal variation in October was also higher than the Val Roseg kryal systems in June and August and the Macun October kryal system (Tukey’s HSD: P<0.05). 295 Ordination of environmental parameters (Figure S1) revealed differences in physicochemical characteristics between water sources, catchments and dates (PERMANOVA: F1,89=21.22, F2,89=24.59 and F2,89=8.89, respectively, P<0.001). An interaction between catchment and dates, and catchment and water source was apparent (PERMANOVA: F3,89=5.04, and F2,89=8.52, P<0.001). Physico-chemical characteristics were not different 300 between Loetschental and Val Roseg krenal waters in August and October (PERMANOVA: F1,13=2.31, P=0.06 and F1,13=3.37, P>0.59, respectively). Macun showed no significant difference between water systems in August or October (PERMANOVA: F1,17=1.33, P=0.24 and F1,8=3.18, P=0.054, respectively). Loetschental krenal systems did not significantly differ from kryal waters in October and June (PERMANOVA: F1,6=0.23, P=0.872, F1,9=1.26, P=0.297, respectively). 305 Loetschental krenal systems did not differ between June and August (PERMANOVA: F1,11=1.67, P=0.147). Val Roseg showed no shift in physico-chemical characteristics between October and June in krenal systems (PERMANOVA: F1,13=.16, P=0.375). 12 310 Detailed results of enzymatic activities The total activity of the enzyme set showed a difference between catchments and water source (ANOVA: F2,89=29.53, P<0.001, F1,89=5.20, P<0.05, respectively). A post-hoc test revealed generally highest mean enzyme expressions in krenal systems (Tukey’s HSD: P<0.05) and highest mean values in Roseg, intermediate values in Loetschental and lowest values in 315 Macun (Tukey’s HSD: P<0.01). There was also a significant interaction between water source and sampling date (ANOVA: F2,89=3.66, P<0.05), Alph had highest activities in krenal systems (ANOVA: F1,89=28.87, P<0.001, Tukey’s HSD: P<0.001) and lowest activities in Macun (ANOVA: F2,89=17.70 P<0.001, Tukey’s HSD: P<0.001). 320 Bet had a significant interaction between catchment, water and sampling date (ANOVA: F3,89=3.64, P<0.05 ), with krenal sites being more active than kryal sites in June and August in Val Roseg (Tukey’s HSD: P<0.05). Roseg had higher Bet activities than the other two catchments (Tukey’s HSD: P<0.05). Xyl had a significant interaction between water, catchment and sampling date, being 325 higher in Roseg krenal sites than in Macun krenal sites in August (ANOVA: F3,89=2.94, P<0.05, Tukey’s HSD: P<0.05). In June, kryal sites in Val Roseg were lower in activities than krenal sites (Tukey’s HSD: P<0.001). There was generally less Xyl activity in krenal sites in Macun than in the other catchments (ANOVA: F2,89=4.98, P<0.01, Tukey’s HSD: P<0.05), and krenal sites were more active than kryal sites in Loetschental (Tukey’s HSD: P<0.05). Roseg and Loetschental 330 had more Xyl activity than Macun (ANOVA: F2,89=6.01, P<0.001, Tukey’s HSD: P<0.05), and krenal sites had more activity then kryal sites (ANOVA: F1,89=43.64, P<0.001, Tukey’s HSD: P<0.001). Est had different activities between the three catchments, being highest in Roseg, intermediate in Loetschental and lowest in Macun (ANOVA: F2,89=85.93, P<0.001, Tukey’s HSD: 335 P<0.005). 13 Nac was lower in Loetschental compared to Val Roseg in August (ANOVA: F3,89=4.61, P<0.05, Tukey’s HSD: P<0.05). In Val Roseg, krenal sites had higher activity than kryal sites, and also higher activity than krenal and kryal sites in Macun and kryal sites in Loetschental (ANOVA: F2,89=5.66, P<0.01, Tukey’s HSD: P<0.001, P<0.01, 340 P<0.05 and P<0.001, respectively). The krenal systems showed generally higher Nac activity (ANOVA: F1,89=33.14, P<0.001, Tukey’s HSD: P<0.001). Leu had different activities between water systems with higher values occurring in krenal sites (ANOVA: F1,89=19.87, P<0.01) and highest activities in Roseg followed by Loetschental and Macun (ANOVA: F2,89=25.24, P<0.001, Tukey’s HSD: P<0.05). 345 End was highest in Val Roseg, intermittent in Loetschental and lowest in Macun (ANOVA: F2,89=65.86, P<0.001, Tukey’s HSD: P<0.001). Kryal systems had the highest End activities (ANOVA: F1,89=14.85, P<0.001, Tukey’s HSD: P<0.001). In Val Roseg krenal sites Phos activity was higher compared to all other water systems within the three catchments except for Macun krenal system which did not differ (ANOVA: 350 F2,89=6.02, P<0.01, Tukey’s HSD: P<0.05). Val Roseg and Macun were generally more active for Phos compared to Loetschental (ANOVA: F2,89=4.73, P<0.05, Tukey’s HSD: P<0.05), and krenal sites showed higher Phos activity then kryal sites (ANOVA: F1,89=48.07, P<0.001, Tukey’s HSD: P<0.001). MHGD revealed a difference in variation of enzymatic activity patterns between 355 groupings of same water source, sampling date and catchment (Permutation test: F15,89=7.13, P<0.001, Figure S3). An interaction of distance to centroids with catchment, water source and sampling date was apparent (Anova: F3,89=2.72, P<0.05). An interaction of distance to centroid with catchment and water source with low variability (Anova: F1,89=15.17, P<0.001), water source and sampling date (Anova: F1,89=4.86, P<0.01) as well as catchment and sampling date 360 (Anova: F1,89=2.86, P<0.05) was apparent. There was also an influence of water source (Anova: F1,89=20.46, P<0.001). In Loetschental and Val Roseg there was low turnover between 14 enzymatic activity patterns within the krenal sites (Tukey’s HSD: P<0.05). Macun had similar enzymatic turnover on the sampled dates in both water systems (Tukey’s HSD: P<0.05).Loetschental kryal sites had much higher turnover in EF in June and August compared 365 to the other sites at any time, except for Loetschental kryal sites in October and Val Roseg krenal sites in August and October and Val Roseg kryal site in June (Tukey’s HSD: P<0.05). The Val Roseg kryal sites in June had higher enzymatic turnover compared to Macun kryal channels during October, Val Roseg kryal channel during August and all krenal channels except for Val Roseg krenal sites in August and October (Tukey’s HSD: P<0.05). 370 Ordination of enzyme activities showed strong separation between the two water systems, catchments and their interactions (Figure 4, Figure S4), (PERMANOVA: F1,89=40.51, P<0.001, F2,89=18.92, P<0.001 and F2,89=5.11, P<0.01, respectively) with clear separation of the three catchments and separation of kryal from krenal in Val Roseg and Loetschental. A temporal shift in EF was partially present in Val Roseg in the pairwise comparison model, although not or 375 only marginally significant in the complete model (PERMANOVA: F2,89=2.02, P=0.08, appendix Table 4). Macun showed no functional separation between water sources (Figures 4 and S4, appendix Table 4). The Macun catchment differed in both water systems from krenal systems of the other catchments in all sampling dates (PERMANOVA: Macun vs. Roseg: F1,48=13.64, P<0.001, Macun vs. Loetschental: F1,41=25.88, P<0.001). Macun water systems were not 380 significantly different in activity patterns (PERMANOVA: F2,26=1,27, P=0.26), whereas the other two catchments showed strong separation in activity structures between krenal and kryal sites (PERMANOVA: Roseg: F1,50=25.85, P<0.001, Loetschental: F1,26=7.42, P<0.01). No differences could be detected between Loetschental and Roseg activities within each water system within any sampling date (appendix Table 1). A temporal shift in enzymatic pattern was present only in 385 Roseg for both water systems (PERMANOVA: F2,28=2.645, P<0.05 and F2,21=2.137, P<0.05, respectively, appendix Table 1). 15 RDA analysis accounted 55.8% of variation in EF to physico-chemical factors (R2adj=0.558, F6,98=22.85, P<0.001). See appendix Table 6 for canonical coefficients of the first two constraint axes and the R2 values of the forward selected environmental parameters (R2 390 were not adjusted, thus values are slightly higher). Variation partitioning revealed that the total contribution of chemical/physical factors on enzymatic activity was 42.6% / 0.3% of explained variation. The shared fraction explained 13.1% of the variation in EF (Total R2adj=0.560, F7,97=19.94, P<0.01, chemical fraction: R2adj=0.426 F6,97=17.65, P<0.01, physical fraction R2adj =0.003, F1,97=1.62, P=0.18, joint fraction R2adj=0.133, not testable). When variation partitioning 395 was assessed for the two water sources independently, there was 48 % of variation in EF explained by chemical factors, 0% by physical factors and 7.4 % by the shared fraction in the kryal systems (Total R2adj=0.553, F6,43=11.09, P<0.01, chemical fraction: R2adj=0.479, F5,43=11.29, P<0.01, physical fraction R2adj =-0.001, F1,43=0.90, P=0.50, joint fraction R2adj=0.074, not testable). The variation partitioning within the krenal system revealed that none of the 400 physical factors were kept in the model after forward selection. Thus, forward selected chemical factors were tested independently and explained 47.9% of the variation of enzymatic activities (Chemical factors: R2adj=0.479, F3,51=17.35, P<0.001). Detailed results of cell abundance 405 Bacteria cell abundance in sediments ranged from 1.66x106 to 4.44x109 (mean: 2.53 x108±6.75x108, n=105) cells per g sediment dry weight (dw) and differed between the three catchments (ANOVA: F2,89=71.45, P<0.001). Loetschental had the lowest mean cell densities (range: 2.65 x106 to 1.90x108 cells g-1 dw, mean: 3.47 x107±4.82 x107 cells g-1 dw, n=27) followed by Val Roseg (range 1.67x106 to 2.75x109 cells g-1 dw, mean: 1.36 x108±4.02 410 x108cells g-1 dw, n=51) and then Macun (range: 5.25 x107 to 4.44x109 cells g-1 dw, mean: 6.92x108±1.11 x109 cells g-1 dw, n=27). Krenal systems had generally higher cell numbers ((ANOVA: F1,89=90.56, P<0.001, Tukey’s HSD: P<0.001). There were higher cell abundances in 16 August than in October (ANOVA: F2,89=3.93, P<0.05, Tukey’s HSD: P<0.05). There was a significant interaction between water source and catchment, with lower cell abundances in kryal 415 sediments in Loetschental and Val Roseg (ANOVA: F2,89=14.45, P<0.001, Tukey’s HSD: P<0.001, appendix Table 1). An interaction of water source and sampling date was also apparent with lowest cell abundance in kryal systems in June and October, intermediate in kryal systems in August and generally higher abundances in krenal systems during all sampling dates with the August samples being significantly higher than the kryal August ones (ANOVA: 420 F2,89=3.56, P<0.05, Tukey’s HSD: P<0.0). Detailed results of bacterial community structure and linked functions The number of all 191 detected OTUs ranged from 28 at site VR12 in June to 127 at site M13 in August. There was a significant interaction between catchment and water source on OTU 425 richness (ANOVA: F2,89=4.19, P<0.05). The number of OTUs differed between water source in Val Roseg with a higher number of OTUs in krenal than kryal sites (Tukey’s HSD: P<0.001). Loetschental had a lower number of OTUs in krenal sites than Val Roseg (Tukey’s HSD: P<0.05) and a lower number of OTUs in kryal sites compared to krenal sites in Macun and Val Roseg (Tukey’s HSD: P<0.05). Diversity (Shannon index) was highest in krenal systems 430 (ANOVA: F1,89=24.19, P<0.001) NMDS ordination showed a differentiation in BCC between the two water sources and the three catchments, but a less pronounced effect of sampling date (PERMANOVA: F1,89=13.21, P<0.001, F2,89=6.36, P<0.001, and F2,89=1.57, P<0.05, respectively, Figure 5 and S6). There was also an interaction between water source and catchment (PERMANOVA: 435 F2,89=4.5, P<0.001). Krenal BCC were similar in Loetschental and Val Roseg in August and June (PERMANOVA: F1,13=0.90, P=0.709, F1,13=1.07, P=0.378, respectively). Otherwise, there was always a significant catchment difference in BCC for the same water source within the same sampling dates (PERMANOVAs: P<0.05, appendix Table 4). Macun was more separated from 17 the other two catchments in the NMDS, and showed a less pronounced separation of kryal and 440 krenal sites, although they still were significantly different (PERMANOVA: F1,26=2.09, P<0.01, Figures 5 and S6). Krenal systems showed no temporal pattern compared to the temporal shift in BCC in kryal sites in Val Roseg (PERMANOVA: F2,21=0.64, P=0.96, F2,28=2.39, P<0.001, respectively). Temporal BCC changes were not significant in Loetschental krenal and kryal sites (PERMANOVA: F2,14=0.70, P=0.896, F2,11=0.82, P=0.610, respectively). Macun BCC showed no 445 temporal pattern in kryal sites but did in krenal sites (PERMANOVA: F1,8=0.77, P=0.791, F1,16=1.75, P<0.05, respectively). MHGD showed a significant difference in dispersion between groupings of the same water source, sampling date and catchment (Permutation test: F15,89=3.53, P<0.001, Figure S3). Distances to centroids differed between water sources (Anova: F1,89=12.53, P<0.001) and 450 sampling date (Anova: F2,89=4.64, P<0.05) Tukey’s HSD revealed lower beta diversity in Val Roseg kryal sites in October compared to all krenal systems except to Macun and Val Roseg krenal systems in October. It was also different to Macun and Val Roseg August kryal systems (Tukey’s HSD: P<0.05). RDA revealed that 19.9% of total variation in BCC was explained by forward selected 455 environmental factors (physico-chemical, R2adj=0.199, F8,96=4.23, P<0.001). See appendix table 7 for canonical coefficients of the first two constraint axes and the cumulative R2 values of the forward selected variables. Variation partitioning showed that the total contributions of chemical/physical factors on BCC were 13.5% / 1.6% of total variation. The shared fraction explained 5.2% of the variation (Total R2adj=0.202, F9,95=3.93, P<0.01, chemical fraction: 460 R2adj=0.135, F8,95=3.17, P<0.01, physical fraction R2adj =0.016, F1,95=2.91, P<0.01, joint fraction R2adj=0.052, not testable). Variation partitioning of physical and chemical parameters applied to the kryal systems showed 23.2% of the variation in community structure was accounted for solely by water chemistry (Total R2adj=0.288, F8,41=3.47, P<0.01, chemical fraction: R2adj=.0.232, F7,41=3.23, P<0.01, physical fraction R2adj =0.014, F1,41=1.81, P<0.05, joint fraction R2adj=0.042, 18 465 not testable). Krenal systems, in contrast, had just 10.9% of the variation in BCC explained by water chemistry (Total R2adj=0.128, F6,48=2.32, P<0.01, chemical fraction: R2adj=0.109, F5.48=2.33, P<0.01, physical fraction R2adj =0.016, F1,48=1.88, P<0.01, joint fraction R2adj=0.003, not testable). Enzyme activities of Phos and Alph were significantly situated towards krenal bacterial 470 communities (fitting significance: P<0.01 and P<0.05 respectively, based on 999 permutations, Figure S6). Gradient directions tended towards Roseg and Loetschental for Alph and towards Macun for Phos. Xyl and Leu also were situated more towards krenal systems, but in a less pronounced manner (fitting significance: P<0.1, based on 999 permutations). Bet and Nac were not fitted significantly in the ordination (P=0.391 and P=0.252, respectively), although they have 475 a weak gradient towards the Macun kryal system. Est and End enzyme activity fitted different from the other tested enzymes with a gradient direction towards kryal systems. Ordination of enzyme activity structure underlines this functional separation between water sources, except for Macun (Figures 4 and S4). The tendency of generally higher Phos and Nac activity was apparent for the Macun catchment. Bet activity centroid, which showed a 480 gradient towards Macun kryal systems in the community structure NMDS, was situated more near the Roseg and Loetschental krenal systems. This result is probably due to the weak environmental fitting power in the community structure ordination. Relating enzyme activity patterns to BCC by means of pairwise comparison of PERMANOVA’s (appendix Table 4) revealed a link between changes in assemblage structure 485 and function in 63.3% of all pairwise comparisons. A low 16.7% of all pairwise comparisons were significant in Macun, indicating a weak linkage between structure and function. Roseg and Loetschental had linkages in 80.0% and 66.7% of all cases, respectively. Procrustes analysis of community and function NMDS showed linkage between BCC and EF (r=0.639, p<0.001) when performed with all catchments included. Correlation of single catchments in the same ordination 490 showed differences in strength of association with maximum correlations for Val Roseg, 19 intermediate correlation for Loetschental, and minimal correlation for Macun. Mantel tests supported these findings. Relating enzyme activity patterns to BCC by means of Mantel tests showed the same trend as the procrustes analysis and the pairwise PERMANOVA comparison: Correlation of 495 structure and function of all catchments was 0.363 (P<0.01). The single catchments showed correlations of 0.561 (P<0.01) in Val Roseg, 0.389 (P<0.01) in Loetschental, and -0.102 (P=0.79) in Macun. Procrustes analysis for community structure and environmental variables were correlated (r=0.601, P<0.01) when performed with all catchments. Correlation of single catchments of the 500 same ordination showed maximum correlations for Val Roseg (r=0.600, P<0.001), minimum for Loetschental (r=0.425, P<0.05) and intermediate for Macun (r=0.521, P<0.01). Correlations between enzymatic structure and environmental structure were for 0.58 for all catchments, (r=0.584 P<0.001), 0.62 for Loetschental (kryal and krenal: r=0.623, P<0.001; kryal: r=0.846, P<0.001; krenal r=0.295, P=0.789), 0.52 for Val Roseg and (kryal and krenal: r=0.519, P<0.001; 505 kryal: r=0.331, P=0.088; krenal r=0.2865, P=0.534) and 0.32 for Macun (kryal and krenal: r=0.320, P=0.149; kryal: r=0.344, P=0.802; krenal r=0.409, P=0.121). See also table 4. Refinement of individual dependency of enzymatic activity on environmental variables by multiple linear regression and relative importance metrics revealed that PP and NO 2-N had a negative influence (or correlation) on most of enzymes, whereas pH, D90D10, PN, PO4-P, 510 alkalinity and temperature positively influenced enzyme activities (Table 5). Fitting single OTU’s on the community function NMDS outcropped 108 OTU’s out of 191 OTU’s that could have been fitted with a permutation power of P<0.05, 74 OTU's with a power <0.01, 50 with power <0.001, 30 OTU's with power <0.0001, and 17 OTU's with a power of <0.00001 (100000 permutations). OTU's with fitting power <0.01 were split into a fraction 515 showing a direction of gradient towards the kryal systems of Val Roseg and Loetschental and a fraction towards the Macun catchment. Only 4 OTU’s showed a gradient towards Roseg and 20 Loetschental krenal system with power P<0.001. One OTU could be fitted with a power below P<0.0001 towards this system (appendix Figure S4). 21 References 520 Anderson MJ. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32-46. 525 Anderson MJ, Ellingsen KE, Mcardle BH. (2006). Multivariate dispersion as a measure of beta diversity. Ecol Lett 9: 683-693. BAFU annual report (2010). Federal Office for the Environment (FOEN): Switzerland. Bennion H, Carvalho L, Sayer CD, Simpson GL, Wischnewski J. (2011). Identifying from recent sediment records the effects of nutrients and climate on diatom dynamics in loch leven. Freshwat Biol 57: 2015-2029. 530 Blanchet FG, Legendre P, Borcard D. (2008). Forward selection of explanatory variables. Ecology 89: 2623-2632. Bray JR, Curtis JT. (1957). An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27: 325-349. 535 Bürgmann H, Jenni S, Vazquez F, Udert KM. (2011). Regime shift and microbial dynamics in a sequencing batch reactor for nitrification and anammox treatment of urine. Appl Environ Microbiol 77: 5897-5907. Chevan A, Sutherland M. (1991). Hierarchical partitioning. Am Stat 45: 90-96. Digby PGN, Kempton RA. (1987). Multivariate analysis of ecological communities. Chapman and Hall: New York. 540 Findlay S, Quinn JM, Hickey CW, Burrell G, Downes M. (2001). Effects of land use and riparian flowpath on delivery of dissolved organic carbon to streams. Limnol Oceanogr 46: 345-355. Gower JC. (1975). Generalized procrustes analysis. Psychometrika 40: 33-51. Grömping U. (2006). Relative importance for linear regression in R: The package relaimpo. J Stat Softw 17: 1-27. 545 Jackson DA. (1995). Protest - a procrustean randomization test of community environment concordance. Ecoscience 2: 297-303. Krambeck HJ. (1995). Application and abuse of statistical methods in mathematical modelling in limnology. Ecol Model 78: 7-15. Labhart TP. (1998). Geology of Switzerland. Geologie der schweiz. Ott Verlag: Thun. 22 550 Maisch M. (1988). The movements of glaciers and snow-lines since the 1850 advance in the canton of Graubunden, Switzerland. Die Veraenderungen der Gletscherflaechen und Schneegrenzen seit dem Hoechstand von 1850 im Kanton Graubunden (Schweiz) 70: 113-130. Malard F, Tockner K, Ward JV. (1999). Shifting dominance of subcatchment water sources and flow paths in a glacial floodplain, val roseg, switzerland. Arct Antarct Alp Res 31: 135-150. 555 Malard F, Tockner K, Ward JV. (2000). Physico-chemical heterogeneity in a glacial riverscape. Landscape Ecol 15: 679-695. Mantel N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Res 27: 209-220. 560 Oksanen J, Blanchet FG, Kindt R, Legendre P, O'hara RB, Simpson GL et al. (2011). Vegan: Community ecology, R package version 1.17-6, http://cran.R-project.Org/package=vegan. Peres-Neto PR, Jackson DA. (2001). How well do multivariate data sets match? The advantages of a procrustean superimposition approach over the mantel test. Oecologia 129: 169-178. Peres-Neto PR, Legendre P, Dray S, Borcard D. (2006). Variation partitioning of species data matrices: Estimation and comparison of fractions. Ecology 87: 2614-2625. 565 Pernthaler J, Glöckner FO, Schönhuber W, Amann R (2001). Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. In: Paul JH (ed). Methods in microbiology. Academic Press. pp 207-226. Porter KG, Feig YS. (1980). The use of dapi for identifying and counting aquatic microflora. Limnol Oceanogr 25: 943-948. 570 R development core team. (2012). R: A language and environment for statistical computing. R foundation for statistical computing. Vienna, Austria. http://www.R-project.org/. Ramette A. (2009). Quantitative community fingerprinting methods for estimating the abundance of operational taxonomic units in natural microbial communities. Appl Environ Microbiol 75: 2495-2505. 575 Robinson CT, Matthaei S. (2007). Hydrological heterogeneity of an alpine stream-lake network in Switzerland. Hydrol Process 21: 3146-3154. Schmidt S, Weber B, Winiger M. (2009). Analyses of seasonal snow disappearance in an alpine valley from micro- to meso-scale (Loetschental, Switzerland). Hydrol Process 23: 1041-1051. 580 Selinummi J, Seppälä J, Yli-Harja O, Puhakka JA. (2005). Software for quantification of labeled bacteria from digital microscope images by automated image analysis. Biotechniques 39: 859862. 23 Tockner K, Malard F, Burgherr P, Robinson CT, Uehlinger U, Zah R et al. (1997). Physicochemical characterization of channel types in a glacial floodplain ecosystem (Val Roseg, Switzerland). Archiv für Hydrobiologie 140: 433-463. 585 Tockner K, Malard F, Uehlinger U, Ward JV. (2002). Nutrients and organic matter in a glacial river-floodplain system (Val Roseg, Switzerland). Limnol Oceanogr 47: 266-277. Yannarell AC, Kent AD, Lauster GH, Kratz TK, Triplett EW. (2003). Temporal patterns in bacterial communities in three temperate lakes of different trophic status. Microb Ecol 46: 391405. 590 24 Supplementary Tables Appendix Table 1 Physico-chemical parameters, given are the average ± standard deviation, N = number of sampled sites Physico-chemical and microbial parameters Catchment 595 Samping Date A Val Roseg O J A Loetschental O J 600 A Macun O Catchment Samping Date A Val Roseg O J A Loetschental O J A Macun O Stream type N Temperature [°C] Conductivity [µS cm -1] pH D90D10 OM [g 100g-1dw] DOC [mg C L-1] POC [mg C L-1] TIC [mg C L-1] kryal krenal kryal krenal kryal krenal 10 8 10 6 9 8 3.12 6.98 2.46 3.65 3.69 6.23 ± ± ± ± ± ± 2.16 1.96 1.73 0.55 2.27 2.64 25.12 44.20 58.97 72.57 102.91 78.54 ± ± ± ± ± ± 1.83 27.96 17.42 22.74 14.87 41.03 7.51 7.00 7.58 6.93 7.48 6.90 ± ± ± ± ± ± 0.34 0.38 0.41 0.30 0.32 0.38 71.99 111.26 48.67 36.48 56.12 156.26 ± ± ± ± ± ± 51.69 126.51 43.22 23.57 74.65 267.12 0.31 0.49 0.35 0.87 0.31 1.10 ± ± ± ± ± ± 0.10 0.22 0.05 0.84 0.09 1.20 0.77 0.39 1.47 1.00 0.37 1.69 ± ± ± ± ± ± 1.66 0.20 1.02 0.53 0.24 3.18 0.36 0.27 1.58 0.16 0.27 0.20 ± ± ± ± ± ± 0.10 0.17 2.60 0.10 0.10 0.05 2.78 5.12 5.79 7.58 5.60 6.51 ± ± ± ± ± ± 0.30 2.80 1.94 1.81 0.99 2.62 kryal krenal kryal krenal kryal krenal 4 6 3 4 4 6 6.20 13.67 5.00 6.43 5.40 9.43 ± ± ± ± ± ± 2.86 5.63 2.03 3.25 1.92 4.63 27.63 76.38 40.25 102.67 66.20 96.88 ± ± ± ± ± ± 6.99 28.97 11.44 87.36 27.84 48.09 6.79 6.51 6.40 6.53 7.05 6.53 ± ± ± ± ± ± 0.35 0.26 0.32 0.27 0.40 0.24 14.41 58.77 57.41 27.81 14.30 97.42 ± ± ± ± ± ± 3.02 65.82 75.49 15.83 2.88 122.04 0.33 0.74 0.88 0.65 0.14 0.51 ± ± ± ± ± ± 0.04 0.33 0.58 0.38 0.04 0.22 0.13 0.48 3.59 1.06 5.70 3.38 ± ± ± ± ± ± 0.04 0.34 3.87 0.61 5.58 5.64 0.42 0.27 0.44 1.89 0.34 0.61 ± ± ± ± ± ± 0.32 0.09 0.36 3.14 0.13 0.60 2.66 4.69 4.12 4.74 3.25 4.33 ± ± ± ± ± ± 0.57 1.54 0.75 1.06 1.00 1.93 kryal krenal kryal krenal 6 11 3 7 7.08 13.75 4.20 4.45 ± ± ± ± 5.13 3.01 0.14 0.90 6.45 7.20 12.00 9.04 ± ± ± ± 3.18 2.85 1.41 3.74 4.80 4.79 4.80 5.03 ± ± ± ± 0.24 0.35 0.09 0.25 63.71 48.10 24.24 44.26 ± ± ± ± 66.58 27.24 19.52 22.97 2.64 3.28 1.14 1.81 ± ± ± ± 2.74 3.72 0.06 1.47 0.45 1.23 1.08 4.98 ± ± ± ± 0.09 1.04 0.18 5.56 0.37 0.92 0.34 1.53 ± ± ± ± 0.26 1.44 0.28 1.12 2.29 2.13 0.83 1.16 ± ± ± ± 1.46 1.37 0.02 0.21 Stream type N NH4-N [µg L-1] NO2-N [µg N L-1] NO3-N [mg N L-1] DN [mg N L-1] PN [mg N L-1] PO4 -P [µg P L-1] DP [µg P L-1] PP [µg P L-1] kryal krenal kryal krenal kryal krenal 10 8 10 6 9 8 22.12 ± 15.28 2.64 ± 0.40 0.13 ± 0.00 0.25 ± 0.00 0.03 ± 0.00 5.29 ± 2.41 5.71 ± 2.83 219.05 ± 81.59 2.50 ± 0.00 1.01 ± 0.04 0.15 ± 0.07 0.25 ± 0.00 0.02 ± 0.02 2.50 ± 0.00 2.50 ± 0.00 10.93 ± 14.28 7.35 ± 5.95 1.89 ± 1.38 0.13 ± 0.00 0.26 ± 0.02 0.06 ± 0.06 7.94 ± 7.02 9.10 ± 8.79 3.02 ± 1.27 1.00 ± 0.00 0.15 ± 0.06 0.26 ± 0.02 0.01 ± 0.01 2.50 ± 0.00 2.97 ± 1.14 11.23 ± 6.76 1.64 ± 0.45 0.22 ± 0.11 0.25 ± 0.00 0.04 ± 0.01 3.21 ± 1.45 4.30 ± 2.19 40.78 ± 37.04 2.50 ± 0.00 1.00 ± 0.00 0.20 ± 0.09 0.25 ± 0.00 0.03 ± 0.02 2.50 ± 0.00 2.50 ± 0.00 0.79 ± 0.54 kryal krenal kryal krenal kryal krenal 4 6 3 4 4 6 6.80 ± 5.28 1.03 ± 0.05 0.13 ± 0.00 0.25 ± 0.00 0.02 ± 0.01 2.50 ± 0.00 2.50 ± 0.00 7.27 ± 6.15 1.00 ± 0.00 0.13 ± 0.00 0.25 ± 0.01 0.02 ± 0.01 2.50 ± 0.00 2.50 ± 0.00 2.50 ± 0.00 1.00 ± 0.00 0.13 ± 0.00 0.25 ± 0.00 0.02 ± 0.01 3.33 ± 1.28 3.25 ± 1.14 62.68 ± 79.16 2.50 ± 0.00 1.00 ± 0.00 0.13 ± 0.00 0.25 ± 0.00 0.17 ± 0.29 2.50 ± 0.00 2.50 ± 0.00 56.79 ± 96.08 20.30 ± 14.07 1.25 ± 0.50 0.13 ± 0.00 0.25 ± 0.00 0.04 ± 0.01 7.05 ± 6.55 8.65 ± 9.65 41.60 ± 52.78 11.35 ± 8.92 1.17 ± 0.41 0.13 ± 0.00 0.25 ± 0.00 0.06 ± 0.05 4.58 ± 2.89 4.77 ± 3.23 14.26 ± 10.24 kryal krenal kryal krenal 6 11 3 7 5.22 ± 4.26 1.00 ± 0.00 0.16 ± 0.03 0.25 ± 0.00 0.03 ± 0.01 6.18 ± 4.80 7.50 ± 5.31 2.45 ± 1.94 9.76 ± 13.44 1.00 ± 0.00 0.13 ± 0.00 0.25 ± 0.00 0.07 ± 0.05 4.39 ± 3.11 6.48 ± 5.11 2.80 ± 2.96 2.50 ± 0.00 1.00 ± 0.00 0.26 ± 0.11 0.39 ± 0.11 0.06 ± 0.04 2.50 ± 0.00 2.50 ± 0.00 6.07 ± 7.16 1.00 ± 0.00 0.13 ± 0.00 0.25 ± 0.00 0.11 ± 0.07 2.50 ± 0.00 2.93 ± 1.06 2.83 ± 3.30 10.13 ± 14.79 25 Bacteria abundance 2.01E+07 ± 1.44E+07 1.20E+08 ± 1.34E+08 454.72 ± 722.43 4.68E+06 ± 2.31E+06 3.12E+08 ± 2.56E+08 1.68 ± 1.49 5.72E+06 ± 4.86E+06 4.73E+08 ± 9.31E+08 154.91 ± 165.75 1.11E+07 ± 6.00E+06 20.77 ± 19.74 5.46E+07 ± 6.80E+07 5.99E+06 ± 2.24E+06 7.72E+07 ± 7.44E+07 9.22E+06 ± 5.88E+06 4.54E+07 ± 4.30E+07 5.84E+08 ± 8.51E+08 1.14E+09 ± 1.46E+09 1.30E+08 ± 3.42E+07 1.85E+08 ± 1.21E+08 Appendix Table 2 Summary of ANOVA analysis for the eight measured enzymes, given are F-ratios, see text for enzyme abbreviations Source of variation df Alph Bet Xyl Est Function Nac Catchment (C) 2 17.69*** 11.13*** 6.01* 85.93*** 0.51 25.24*** 65.86*** 4.73* 32.51*** Watersystem (W) 1 28.87*** 28.77*** 43.64*** 3.10 33.15*** 19.87*** 14.85*** 48.07*** 17.43*** Sampling Date (S) 2 1.35 2.08 1.71 1.38 0.85 1.29 1.51 1.21 1.45 CxW 2 2.37 3.33* 4.98** 1.96 5.66** 1.94 2.84 6.02** 1.60 CxS 3 0.68 1.94 2.72* 1.15 4.61** 2.04 0.41 1.94 2.18 WxS 2 0.19 0.06 0.17 1.00 1.84 2.11 0.76 0.80 0.92 CxW xS 3 1.56 3.64* 2.94* 0.29 1.26 1.24 0.55 1.16 1.67 * P<0.05 ** P<0.01 *** P<0.001 605 26 Leu End Phos total activity 610 615 Appendix Table 3 Enzymatic activities (average±SD), N = number of sampled sites, n= 3 replicates per site, see main text for abbreviations Enzymatic activities [n mol substrate g-1 h-1] 620 Catchment Sampling Stream type Date A 625 Val Roseg O J A Loetschental O J A Macun O N kryal krenal kryal krenal kryal krenal 10 8 10 6 9 8 kryal krenal kryal krenal kryal krenal 4 6 3 4 4 6 kryal krenal kryal krenal 6 11 3 7 Alph Bet Xyl Nac Est Leu End 24.11 ± 14.56 85.36 ± 64.65 194.75 ± 260.11 629.57 ± 1045.99 Phos 10.31 ± 6.37 27.18 ± 19.89 679.99 ± 530.18 1091.97 ± 827.63 880.27 ± 953.79 80.25 ± 41.35 54.42 ± 58.97 213.43 ± 292.18 2482.50 ± 1652.01 1511.02 ± 1895.75 269.63 ± 476.81 561.26 ± 782.69 1811.76 ± 1965.82 88.42 ± 134.36 390.61 ± 287.01 23.75 ± 15.44 108.41 ± 52.80 6.77 ± 5.68 9.69 ± 14.22 955.08 ± 2076.65 562.01 ± 1121.61 101.72 ± 107.00 190.64 ± 138.20 18.99 ± 6.17 58.16 ± 38.16 312.95 ± 284.42 5692.19 ± 6424.34 184.59 ± 174.02 56.08 ± 79.42 94.39 ± 87.16 5.57 ± 5.18 16.43 ± 26.58 839.19 ± 909.04 750.85 ± 852.35 1342.77 ± 1332.85 89.84 ± 102.64 141.64 ± 97.72 1333.22 ± 2927.52 114.14 ± 198.01 330.36 ± 777.92 354.15 ± 364.27 3439.80 ± 3840.35 179.37 ± 210.42 914.21 ± 1756.86 9.62 ± 6.43 14.56 ± 15.53 2.52 ± 2.24 11.41 ± 19.24 371.30 ± 523.01 215.43 ± 90.65 357.01 ± 412.59 45.31 ± 12.96 71.58 ± 68.39 153.83 ± 120.23 26.32 ± 30.07 34.73 ± 41.15 109.92 ± 98.30 524.60 ± 392.76 64.03 ± 66.92 153.34 ± 126.01 7.70 ± 4.02 14.59 ± 6.46 3.38 ± 2.19 5.93 ± 5.24 219.38 ± 263.20 191.66 ± 92.50 168.47 ± 204.89 19.83 ± 7.79 91.32 ± 74.85 215.83 ± 227.00 38.00 ± 39.87 104.15 ± 71.68 104.10 ± 86.64 1841.93 ± 2358.75 47.08 ± 40.37 325.85 ± 328.77 44.96 ± 24.29 197.70 ± 173.35 24.48 ± 14.46 73.88 ± 103.59 4307.61 ± 7921.58 1201.03 ± 669.86 2967.09 ± 5375.57 139.74 ± 67.23 114.60 ± 145.52 385.09 ± 571.94 57.60 ± 76.13 124.53 ± 157.96 205.81 ± 281.25 1696.96 ± 2981.65 109.16 ± 143.18 269.46 ± 307.60 9.45 ± 8.06 72.82 ± 75.30 7.21 ± 5.43 44.40 ± 59.93 9.37 ± 3.74 241.19 ± 208.81 50.30 ± 44.86 9.12 ± 9.18 22.03 ± 19.73 6.80 ± 2.88 10.02 ± 10.49 181.24 ± 131.34 10.09 ± 10.67 298.79 ± 272.60 4.08 ± 6.12 279.09 ± 195.58 5.40 ± 3.45 27.92 ± 15.91 3.58 ± 2.34 16.26 ± 12.23 4.74 ± 0.65 111.82 ± 54.57 3.01 ± 5.14 79.66 ± 35.70 8.83 ± 10.53 52.07 ± 51.89 12.21 ± 5.74 553.61 ± 383.06 1.82 ± 2.53 404.74 ± 278.47 26.28 ± 36.88 114.85 ± 171.65 27 Appendix Table 4 F-Values of pairwise PERMANOVA comparisons of community structure and enzymatic activities. Groups are split by catchments, water source and season, dF tot are given in parentheses Catchment 630 Season Catchment Season Summer krenal kryal Macun 635 Water Winter krenal kryal Summer krenal kryal 640 Val Roseg Winter krenal kryal Spring krenal kryal Summer 645 krenal kryal Loetschental Winter krenal kryal Spring krenal kryal Macun Summer Val Roseg Winter Summer ARISA (17)0.89 (17)0.99 (14)0.82 (19)3.11* (21)41.96*** (17)9.13*** (21)31.67***(19)10.48***(20)27.84*** (11)1.55 (8)0.68 (13)2.18 (15)29.24*** (11)6.7** (15)19.41*** (13)7.08** (14)14.72** (11)8.08** (9)4.86** (8)3.71* (9)6.03** (8)1.04 (13)2.05 (15)39.43*** (11)5.89** (15)22.74*** (13)8.30** (14)17.29*** (11)13.13** (9)5.75** (8)4.75* (9)7.75** (11)16.44** (14)1.52* (8)0.77 (8)1.79* (10)0.85 (10)1.75* (21)7.41*** (15)4.50*** (15)5.16** (12)3.43** (17)2.37*** (11)1.90** (11)2.20** (8)2.04* (21)11.75*** (15)7.91*** (15)8.56*** (12)6.51** (19)2.78*** (13)2.29*** (13)2.26*** (10)1.96** (20)9.14*** (14)5.98*** (14)6.25** (11)4.86** (17)2.47*** (11)1.68** (11)1.98** (12)25.02** (8)3.91* krenal (12)11.48** (10)3.88* (17)14.55*** (13)2.48* (17)11.51*** (15)1.17 (17)3.98*** (13)0.63 (15)4.63*** (15)5.85** (17)4.59*** (13)0.78 (16)4.61*** (18)2.77** (14)5.71*** (13)2.85* (18)1.56 (16)5.32*** (13)0.90 (15)2.87*** (11)1.32 (15)4.76*** (13)1.13 krenal kryal (5)3.33 (6)3.75 (8)9.64* (6)3.54* (10)1.09 (11)2.50* (13)2.12 (11)2.52 (12)8.71** (13)0.87 (8)2.33 (9)1.57 (14)7.54* (15)20.42** (13)0.60 (11)4.24** (9)1.95 (15)10.10*** (13)1.29 (12)9.28** (10)0.82 (11)3.00* (14)8.58** (12)0.89 (11)6.87* (12)1.86 (9)2.10 (8)2.32 (9)2.54 (11)1.14 (9)2.01 (6)2.00 (7)0.18 (9)3.16 (7)0.13 (6)2.44 (8)1.80 (6)1.71 (9)4.29* (7)0.08 (9)2.03* Enzymes (14)3.48** (13)3.19** (11)2.40*** (12)2.53* (9)1.69* (14)2.41** (8)1.73* (8)1.81* (5)1.55 (10)1.19 (12)1.65 (8)1.52* (12)2.44* (11)1.85 (8)0.83 (6)1.31 (15)4.60*** (9)3.30** (9)3.72** (6)3.06* (11)2.28* (13)2.87** (9)3.20** (13)3.32** (11)2.59*** (12)2.62* (9)2.11* (7)0.61 (6)1.58 (13)0.91 (15)3.37** (11)1.20 (15)6.11*** (14)4.25*** (11)0.56 (9)1.90* (8)0.75 (9)2.61** (11)2.24** (13)3.15** (9)3.5** (13)4.86*** (11)2.52*** (12)3.36** (9)2.01* (7)0.82 (6)1.43 (7)1.06 (8)1.42 (6)3.54* * P<0.05 ** P<0.01 *** P<0.001 28 (13)1.07 (9)1.59 (11)3.34 (9)2.88** (1)1.71* (11)5.90** (13)1.01 (13)1.85* (9)3.24** (15)35.46** (13)0.094 (13)1.23 (15)15.99*** (13)0.80 (11)1.94* (11)1.45* (9)5.37* (6)2.52 (6)2.54* (9)3.12*** (9)4.26** (11)2.62 (9)3.19** (15)4.27** (11)8.56** (13)1.95 (9)2.84** (17)1.98** kryal (17)12.57*** (15)8.98*** (14)5.38** (15)9.84** (17)2.79*** (15)7.85*** (15)4.17*** (10)1.45* krenal (8)5.90** (16)12.92** (8)1.52* kryal (11)8.16* (17)12.48*** (18)2.91 (17)7.44*** krenal (16)11.17** (15)9.38*** (19)2.72* (17)21.34*** (18)2.27 (17)6.47*** (19)2.71** (15)7.87*** (15)0.54 kryal Spring kryal (19)2.54*** (13)2.04*** (13)2.23** kryal Winter krenal (11)1.97** krenal Summer kryal (17)1.75* kryal Loetschental Spring krenal (17)1.41* krenal Winter (13)0.80 (11)2.83 (14)11.58** (12)1.09 (9)2.84 Appendix Table 5 Model parameters of AIC selected multiple linear regression between enzymatic activity and physicochemical parameters and its relative importance metrics. Models are based on enzyme activities standardized to OM 650 Enzyme 655 660 Relative importance metrics Multiple linear regression Alph Bet Xyl Nac Est Leu End Phos AIC modell parameters 8 7 7 6 7 7 7 8 dF Residual SE multiple R2adj F-stat P-Value 96 97 97 98 97 97 97 96 1.007 1.022 0.933 1.236 0.936 1.082 1.213 1.11 0.442 0.444 0.371 0.218 0.757 0.411 0.75 0.356 11.3 12.84 9.76 5.825 47.21 11.37 45.59 8.196 <0.001 <0.001 <0.001 <0.001 <0.001 <0.001 <0.001 <0.001 29 Total responce variance 1.818 1.877 1.384 1.952 3.597 1.987 5.89 1.916 Proportion explained by model 48.49% 48.10% 41.33% 26.29% 77.31% 45.08% 76.69% 40.58% Appendix Table 6 Left side: explained deviance (Dev) of the response surface (generalized additive models) of the 2 environmental variables and fitted factors with their squared correlation coefficients (r ) on the EF NMDS. Right side: Canonical coefficients from the of forward selected physico-chemical parameters incorporated into the global RDA model 2 explaining EF. Given are the first two constraint axes. Explained variations of the constraint axes and R of selected model 2 parameters are based on unadjusted R RDA canonical coefficients NMDS Variables dF F Dev P Temperature Conductivity pH D90D10 OM DOC POC TIC NH4-N NO2-N NO3-N DON PN PO4-P DP PP Factors 5.21 6.22 8.64 2.45 7.17 2.84 6.25 4.20 2.00 6.98 7.96 2.00 2.00 7.08 6.79 6.96 6.55 7.22 32.55 0.17 4.69 0.23 1.07 5.14 12.16 8.22 2.37 0.39 1.30 3.06 2.65 4.39 26.8% 32.5% 74.8% 0.8% <0.001 <0.001 <0.001 0.88 <0.001 0.87 0.39 <0.001 <0.001 <0.001 <0.05 0.68 0.28 <0.01 <0.05 <0.001 P 27.9% 1.2% 8.9% 18.5% 19.3% 38.7% 19.7% 0.8% 2.5% 20.5% 17.9% Catchment 25.7% r2 0.31 Sampling Date 0.07 <0.05 Water 0.26 <0.001 <0.001 30 R2 of forward selected variables RDA1 (52.52%) RDA2 (3.80%) R -0.08 0.44 0.37 0.08 -0.11 0.10 0.03 0.04 0.07 0.07 -0.06 -0.04 0.27 -0.11 0.01 0.01 0.08 -0.06 2 Appendix Table 7 Left side: explained deviance (Dev) of the response surface (generalized additive models) of the environmental variables and fitted factors with their squared correlation coefficients (r 2) on the BCC NMDS. Right side: Canonical coefficients from the of forward selected physico-chemical parameters incorporated into the global RDA model 2 explaining BCC. Given are the first two constraint axes. Explained variations of the constraint axes and R of selected model 2 parameters are based on unadjusted R RDA canonical coefficients NMDS Variables dF F Dev P Temperature Conductivity pH D90D10 OM DOC POC TIC NH4-N NO2-N NO3-N DON PN PO4-P DP PP Factors 6.06 7.28 8.29 2.00 8.58 4.04 6.81 6.90 7.34 5.01 6.53 2.00 5.52 7.81 8.28 6.82 8.22 3.95 17.21 0.50 10.74 1.23 1.34 1.93 4.22 9.15 1.39 2.56 1.65 2.88 3.65 3.50 34.9% 25.8% 60.4% 1.0% 50.5% 6.7% 11.1% 14.4% 26.6% 32.6% 10.7% 4.8% 10.3% 23.1% 28.0% <0.001 <0.001 <0.001 0.61 <0.001 0.30 0.24 0.07 <0.001 <0.001 0.22 0.08 0.15 <0.01 <0.001 <0.01 P Catchment 22.0% r2 0.27 Samling Date 0.06 <0.05 Water 0.30 <0.001 <0.001 31 R2 of forward selected variables RDA1 (14.7%) RDA2 (3.7%) R -0.04 0.01 -0.01 -0.06 -0.02 -0.41 0.02 0.02 0.10 -0.03 0.04 0.01 -0.07 0.04 -0.04 -0.01 0.01 0.02 0.08 0.13 0.05 0.03 0.01 0.02 2 Appendix Table 8 Procrustes analysis of community-, functional- and physico-chemical characteristics based NMDS. Given are r-values (correlations) of the symmetric procrustes rotation, EF = enzymatic functioning, Env = physico-chemical parameters, BCC = bacterial community composition Val Roseg Loetschental Macun Total Total Total EF Env EF Env EF Env BCC 0.668*** 0.600*** 0.551*** 0.425* 0.431* 0.521** kryal krenal kryal krenal kryal krenal EF Env EF Env EF Env EF Env EF Env EF Env BCC 0.558*** 0.375* 0.332 0.482* 0.84*** 0.695** 0.43 0.51 0.358 0.538 0.506* 0.606*** 665 * P<0.05 ** P<0.01 *** P<0.001 32 Appendix Figure S1 NMDS of physico-chemical parameters. Dots indicate individual sites. Coding is equal to sites in figure 1 in the main manuscript, suffix letter indicate sampling date (August: A, October: O, June: 670 J). Dark grey dots correspond to kryal sites and light grey dots to krenal sites. Dispersionellipses depict the standard error of weighted average scores of water source within catchment grouping (confidence limit=0.95). Appendix Figure S2 Boxplots of environmental variables. Whiskers indicate 1.5x interquartile range. Groups are split 675 by water source (brackets), catchments and sampling dates. Appendix Figure S3 Boxplots of multivariate homogeneity of community and enzymatic structure and physicochemical characteristics. Boxes summarize the distances to group centroids on the first two PCoA axes as assessed by MHGD. Whiskers indicate 1.5x interquartile range. Upper panel 680 shows group distances of physico-chemical characteristics, middle panel distances of the community structure (ARISA), and lower panel the distances of bacteria function (Enzymes). Groups are split by water source, catchments and sampling dates Letters show significant differences based on Tukey’s HSD (P<0.05). 685 Appendix Figure S4 NMDS of enzymatic activities. Upper panel: Dots indicate individual sites. The size of dots is relative to the sum of logarithms of all measured enzymes standardized to OM. Orange dots correspond to kryal sites and yellow dots to krenal sites. Dispersion-ellipses depict the standard error of weighted average scores of catchment groupings (confidence limits = 0.95). 690 Environmental variables and bacterial OTU’s are fitted as arrows. Projections of sites on fitted environmental vectors (arrows) show maximum correlation with the corresponding variable and vector lengths indicate strengths of gradient. Significantly fitted vectors are indicated by blue 33 arrows for environmental variables (P<0.05 = dark blue, P<0.05 = light blue) and grey arrows for bacterial OTU's (P<0.001). Lower panel: Dots depicts individual sites. Coding is equal to sites 695 in figure 1 in the main manuscript, suffix letter indicate sampling date (August: A, October: O, June: J). Appendix Figure S5a to S5c NMDS of enzymatic activities with response surface as assessed by generalized additive models (GAM) for the measured physico-chemical variables. The size of dots is relative to the 700 number of OTU’s at a site. Each contour line is annotated with the specific value of the variable. The percentages of variances explained by variables are given and correspond to the values from appendix table 6. Appendix Figure S6 NMDS of ARISA profiles. Upper panel: Dots indicate individual sites. The size of dots is relative 705 to the number of OTU’s at a site. Bright blue dots correspond to kryal sites and dark blue dots to krenal sites. Dispersion-ellipses depict the standard error of weighted average scores of water source (kryal, krenal) within catchment groupings (Macun = M, Loetschental = L, Val Roseg = VR) (confidence limits = 0.95). Environmental variables and enzyme activity are fitted as arrows. Projections of sites on fitted environmental vectors (arrows) show maximum correlation with 710 corresponding variable and vector lengths indicate strengths of gradient. Significantly fitted vectors (P<0.05) are indicated by blue arrows for environmental variables, orange for enzymatic activities and red for bacterial domain and groups. Lower panel: Dots depict individual sites. Coding is equal to sites in figure 1 in the main manuscript, suffix letter indicate sampling date (August: A, October: O, June: J). 715 Appendix Figure S7a to S7c NMDS of ARISA profiles with response surface as assessed by generalized additive models (GAM) for the measured physico-chemical variables. The size of dots is relative to the number of 34 OTU’s at a site. Each contour line is annotated with the specific value of the variable. The 720 percentages of variances explained by variables are given and correspond to the values from appendix table 7. Appendix Figure S8a to S8p Bubble maps of physico-chemical variables. Sizes of the bubbles are standardized by the margin maximum value of distinct variable. Grey dots correspond to krenal sites and green dots 725 to kryal sites. Coding is equal to sites in figure 1 in the main manuscript, suffix letter indicate sampling date (August: A, October: O, June: J). Appendix Figure S9 a to S9c PcoA of (a) physico-chemical, (b) enzymatic activities and (c) community structure. Colors depict the different sites split by catchment, water sources and sampling date. Coding is equal to sites 730 in figure 1 in the main manuscript, suffix letter indicate sampling date (August: A, October: O, June: J). MHGD analysis (i.e. distances to centroids) were based on these ordinations. 35 Appendix Figure S1 735 740 0.4 745 kryal krenal Stress: 12 PN POC 750 M2O M13A M1A M9O 0.2 M16A NMDS2 755 VR13O VR10A VR9A 0.0 PP VR11O -0.2 VR14O VR12A VR13A VR14A VR17A M5A M15A DP NO2-N PO4-P NH4-N L7J OM M14A M17A M8A M krenal M7A M7O M10A M3A M9A M8O M6O M3O DOC M6A M kryal M11A M2A M4A M14O M10O M12A M4O Temp D90D10 VR7A L7O VR2A VR15A VR3A L8J L7AVR18A VR5A VR11A VR12O DON VR10O M12O L kryal L9A L8AL10A L4O VR18O VR kryal L6A VR8A L10O pH L2J VR18J L4A L5J L10J VR15J VR12J VR6A NO3-N L6O L3J L5A L9O VR9O L1J L krenal VR16AL9J L3A L4J VR16O L2A VR5J VR7O VR VR5OVR16J krenal VR1A VR14J VR13J VR15O L5OVR8O VR17J VR10J VR1O VR6J VR3J VR9J VR7J L1A VR2J VR2O L6J VR1J VR8J VR6O TIC VR4O VR4A Conductivity L1O -0.4 -0.2 0.0 NMDS1 36 VR4J 0.2 Appendix Figure S2 0.45 1 DON [mg N L ] 1 DOC [mg C L ] 15 10 5 0 0.35 0.30 0.25 L M VR L M VR L M VR L M VR L M VR L M VR L M VR L M VR 0.5 0.4 7 4 0.30 0.25 1 1 PN [mg N L ] 2.5 POC [mg C L ] 0.40 2.0 1.5 1.0 0.5 0.20 0.15 0.10 0.05 0.0 0.00 L M VR L M VR 400 300 8 D90D10 1 TIC [mg C L ] 10 6 200 4 100 2 0 L M VR L M VR kryal 1 NO3-N [mg N L ] 0.35 0.30 0.25 0.20 0.15 L M kryal VR L M VR krenal 37 krenal Appendix Figure S3 Distance to centroid 0.20 Sampling Month A O J 0.15 a ac ab ab bc ab ab 0.10 Physicochem ical param eters ab bc ab bc bc bc bc b b 0.05 c c Distance to centroid 0.30 ac bc 0.25 ac EF ac ab ab a a 0.20 a a 0.15 a ab a a 0.10 0.05 0.55 a Distance to centroid 760 0.50 0.45 a a a a a ab ab BCC ab a ab 0.40 a ab ab ab b 0.35 0.30 0.25 L M VR kryal L M krenal 38 VR Appendix Figure S4 765 0.6 kryal krenal Stress: 6.64 pH 770 CondX905 0.4 X1085 0.2 775 X1031 X1013 X269 X1121 X695 X1043 X1007 X953 NO3-N PP 780 NMDS2 0.0 X305 X659 X551 X701 X413 Alph DON X923 X881 X1067 X911 X533 X569 TIC X995 X1091 X599 X1175 VR kryal L kryal X509 L krenal Bet Xyl Leu VR krenal PO4-P X809 Est End X941 NO2-N X983 X1073 0.2 X989 DP Nac POC DOC D90D10 PN M kryal X485 X647 M X443 krenal X719 X521 X899 NH4-N X563 X707 OM X605 X203 X1079 X1127 X257 X665 X689 X239 X215 X965 X1283 X455 Phos X425X641 X629 X587 X473 X713 X251 X329 X263 X1187 X419 X347 X317 X851 0.4 Temp X209 X515X575 X1049 785 X773 X407 0.6 X449 X653 0.8 790 kryal krenal Stress: -1.0 -0.5 0.0 0.5 0.4 0.2 NMDS2 0.0 VR14J VR12J 0.2 L6J VR13J L6A VR8J L1O VR6O VR18A VR17J L8AL2AVR3J VR18J VR13O VR2J L8J VR6JL9A L9J VR2O VR6A VR18O VR7O VR9O L7J L3J L5J L10J VR12O VR14O L1J VR17AL10O VR15A L1A VR5J VR9J VR4J L3A L5A L10A VR10O L9OL2JL7AL7O VR10J VR10A L4A VR8A VR13A VR7A VR15JL5O VR3A VR11O VR4A M3O VR16OM11A VR14A L4O VR9AVR5A M4O M13A VR4O M17A M9O VR15O L6O VR1J M10O M12A VR5O VR16J VR12A M12O M15A M5A VR11A VR7J L4J M1A VR1O M3A M14O M16A M8A M4A M7O VR2A VR8O M2A M8O M7A M14A M9A 0.4 M2O VR1A M6O VR16A M6A M10A 0.6 0.8 -1.0 -0.5 0.0 NMDS1 39 0.5 Appendix Figure S5a 1 0.6 Conductivity [ S cm ] Sediment origin 26.8 % Sediment origin 32.5 % kryal krenal 80 0.2 0.4 kryal krenal 0.2 0.4 0.6 Temperature [°C] 70Loetsch.Ground 60 8 6 30 20 0.0 Macun.Glacial Macun.Ground 10 -0.6 -0.6 0 7 5 4 50 -0.4 9 -0.2 Macun.Glacial Macun.Ground 40 Roseg.Ground Roseg.Glacial Loetsch.Glacial 80 3 NMDS2 Roseg.Ground -0.2 0.0 Roseg.Glacial Loetsch.Glacial -0.4 NMDS2 Loetsch.Ground -0.5 0.0 -1.0 -0.5 0.0 NMDS1 pH D90D10 0.6 NMDS1 Sediment origin 74.8 % 0.4 kryal krenal Sediment origin 0.8 % kryal krenal 55 0.2 7 0.2 0.4 0.6 -1.0 Stress: 6.64 -0.8 -0.8 Stress: 6.64 Loetsch.Ground Loetsch.Ground 60 6 Macun.Glacial Macun.Ground 7 Roseg.Ground Roseg.Glacial Loetsch.Glacial 0.0 5 4. 5 5. 5 7. 5 NMDS2 Roseg.Ground 65 Macun.Glacial Macun.Ground 70 -0.2 0.0 Roseg.Glacial Loetsch.Glacial -0.2 NMDS2 6 .5 -0.4 -0.6 -0.6 -0.4 75 -0.5 0.0 -1.0 -0.5 NMDS1 1 1 0.6 DOC [mg C L ] Sediment origin 27.9 % 5 Loetsch.Ground 0.0 1.4 1. 5 1.45 Macun.Glacial Macun.Ground 1. 1. 1. 7 -0.6 -0.6 1. 1.65 85 -0.4 -0.4 2 75 1. 8 1.55 5 1.6 1. 9 NMDS2 3 -0.2 Macun.Glacial Macun.Ground Roseg.Ground Roseg.Glacial Loetsch.Glacial -0.2 2. 5 0.0 Roseg.Ground 1.2 % kryal krenal 1. Loetsch.Ground 0.5 1 Roseg.Glacial Loetsch.Glacial Sediment origin 0.2 0.4 kryal krenal 0.2 0.4 0.6 Organic matter [g g dw] -1.0 -0.5 Stress: 6.64 -0.8 Stress: 6.64 -0.8 NMDS2 0.0 NMDS1 9 -1.0 Stress: 6.64 -0.8 -0.8 Stress: 6.64 0.0 -1.0 NMDS1 -0.5 0.0 NMDS1 40 Appendix Figure S5b 1 1 0.6 TIC [mg C L ] Sediment origin 8.9 % Sediment origin 18.5 % kryal krenal 5.5 0.2 0.4 kryal krenal 0.2 0.4 0.6 POC [mg C L ] 0.4 Loetsch.Ground 1 0. 8 NMDS2 Macun.Glacial Macun.Ground 0. 6 0.9 7 4.5 Macun.Glacial Macun.Ground 4 -0.4 3. 5 -0.5 0.0 -1.0 -0.5 2 0.0 NMDS1 1 1 NO3-N [mg N L ] 0.6 NH4-N [ g L ] Sediment origin 19.3 % 0.2 0.4 kryal krenal Sediment origin 15.9 % kryal krenal 0. 12 0.2 0.6 5 Stress: 6.64 NMDS1 Loetsch.Ground Roseg.Glacial Loetsch.Glacial 0.0 4 Roseg.Ground Macun.Glacial Macun.Ground 0. 0.16 0. 2 6 Macun.Glacial Macun.Ground NMDS2 Roseg.Ground 20 -0.2 Loetsch.Ground 0.14 -0.2 Roseg.Glacial Loetsch.Glacial 16 0.0 12 14 18 NMDS2 2. -0.8 -0.8 Stress: 6.64 -1.0 0.4 3 -0.6 -0.6 0.8 -0.4 0. 5 0. 7 0.0 0.7 1 0. Roseg.Ground Roseg.Glacial Loetsch.Glacial -0.2 0. 3 Loetsch.Ground 5 0. 9 0. 5 Roseg.Ground 0.6 0.0 1.1 -0.2 NMDS2 Roseg.Glacial Loetsch.Glacial 0. 1 -0.4 -0.4 14 8 -0.6 -0.6 10 -1.0 -0.5 0.0 -1.0 NMDS1 1 38.7 % kryal krenal 0.6 0.2 0.4 Sediment origin 0.0 1. 5 1. 3 4 1. 2 1. 6 1. -0.4 Macun.Glacial Macun.Ground -0.6 NMDS2 -0.2 Roseg.Ground 1.9 1. 7 1 1.1 Loetsch.Ground Roseg.Glacial Loetsch.Glacial 1.8 Stress: 6.64 -1.0 -0.5 -0.5 0.0 NMDS1 NO2-N [mg N L ] 2 Stress: 6.64 -0.8 -0.8 Stress: 6.64 -0.8 795 0.0 NMDS1 41 Appendix Figure S5c 800 1 1 0.6 Sediment origin 0.8 % 0.4 kryal krenal 0.0 54 Loetsch.Ground55 0. 2 0. 0 Roseg.Ground 0. 2 0. 2 56 57 58 5 0. 2 0. 2 9 6 0. 0 0. 0 -0.5 0.0 -1.0 5 0. 0 55 0. 0 -0.5 6 1 0.6 Sediment origin 20.5 % 0.4 kryal krenal Sediment origin 17.9 % kryal krenal 0.0 6 7 5 4. Macun.Glacial Macun.Ground -0.4 3 -0.6 -0.6 2 4 -0.4 5 5 6. 5 3. 4 7. 5 5. 8.5 8 4. 5 5 2.5 5 -0.2 Macun.Glacial Macun.Ground 5 Roseg.Ground Roseg.Glacial Loetsch.Glacial 9 5. 5 5. 6 4 6.5 NMDS2 Roseg.Ground Roseg.Glacial Loetsch.Glacial -0.2 0.0 Loetsch.Ground 4.5 NMDS2 7. 5 3.5 4 5 0.2 0.2 3. -0.5 0.0 -1.0 NMDS1 1 Sediment origin 25.7 % 0.2 kryal krenal 0.0 Loetsch.Ground Loetsch.Glacial 50Roseg.Glacial Macun.Glacial Macun.Ground 0 300 Roseg.Ground 15 -0.2 250 0 -0.6 50 -0.4 10 0 200 -0.8 Stress: 6.64 -1.0 -0.5 -0.5 0.0 NMDS1 PP [ g P L ] 3 Stress: 6.64 -0.8 -0.8 Stress: 6.64 -1.0 0.6 7 1 7 0.4 0. 0 DP [ g P L ] 5 NMDS2 65 0.0 Loetsch.Ground 3 825 0. 0 NMDS1 PO4-P [ g P L ] 0.6 Macun.Glacial Macun.Ground Stress: 6.64 NMDS1 0.4 45 4. 5 -1.0 820 Roseg.Ground 4 -0.8 Stress: 6.64 -0.8 815 0. 0 -0.6 -0.6 61 0. 2 Loetsch.Ground 35 Roseg.Glacial Loetsch.Glacial -0.4 -0.2 .2 Macun.Glacial 0Macun.Ground -0.4 810 NMDS2 0. 2 Roseg.Glacial Loetsch.Glacial 53 2.5 % kryal krenal 0.2 0. 2 Sediment origin 0.0 52 NMDS2 0. 2 0.2 805 PN [mg N L ] -0.2 0.4 0.6 DN [mg N L ] 0.0 NMDS1 42 Appendix Figure S6 830 kryal krenal Stress: 15 NO2-N OM 0.5 NH4-N 835 DP PO4-P PP DON M kryal POC Bet Nac D90D10 NO3.N VR kryal Est 0.0 840 NMDS2 End PN Cyt.Flav L kryal TIC 845 Phos M krenal L krenal VR krenal DOC Xyl Leu 0.5 Cond Temp pH Alph 850 -1.0 -0.5 0.5 VR11A 0.0 0.5 L1A M14O L6A L3J L3A M16A M17A M5A M1A VR10A VR12A VR7A M10O M14A VR5A M9O M9A VR1J VR11O VR12O M11A VR1O VR4O VR14A L4A M2A M12A M15A VR9O VR13A M12O VR10O VR4A VR9A L1J VR8A VR10J L4O VR16J M2OVR16O VR14J VR1A VR14O VR13O VR4J M13A VR8J VR13J L5A VR12J M7O VR18JL7J VR16A VR7O VR5O M7A L5J VR18O L1O VR5J M8A L7A M3A VR2O L6O VR6O VR15J VR6A L6J VR8O VR9J L7O M6A VR7J M6O VR2J VR15O M3O VR17A VR3A VR15A L8A L4J L8J VR2A L5O VR18A VR6J L9J M4O VR17J L9O L10O M8O L10A L10J L2J VR3J L2A L9A M4A 0.0 0.5 NMDS2 M10A -1.0 -0.5 0.0 NMDS1 43 0.5 Appendix Figure S7a 855 Temperature [°C] 1 Conductivity [ S cm ] Sediment origin Sediment origin 34.9 % 25.8 % kryal krenal 40 50 Macun.Glacial Loetsch.Glacial 11 7 9 -0.5 8 -0.5 0.0 NMDS2 12 6 70 Loetsch.Glacial 5 Macun.Ground Loetsch.Ground Roseg.Ground 10 0.0 Stress: 15 -1.0 -0.5 0.0 Stress: 15 0.5 -1.0 0.0 NMDS1 pH D90D10 Sediment origin 0.5 Sediment origin 60.4 % kryal krenal 1% kryal krenal 5 0.5 6. 7 0.5 -0.5 NMDS1 5 Macun.Glacial 6 4. Macun.Glacial 5 0.0 Macun.Ground Loetsch.Ground Roseg.Ground 75 Roseg.Glacial 0.0 NMDS2 5 Macun.Ground Loetsch.Ground Roseg.Ground Loetsch.Glacial Loetsch.Glacial 60 50 -0.5 -0.5 45 Stress: 15 -0.5 0.0 Stress: 15 0.5 -1.0 -0.5 0.0 NMDS1 1 1 Organic matter [g g dw] POC [mg C L ] Sediment origin Sediment origin 50.5 % 11.1 % 0.5 kryal krenal 1 3. Macun.Glacial 5 2 3 5 0.8 2. 5 0.5 0 Macun.Ground Loetsch.Ground Roseg.Ground Roseg.Glacial 0.8 0.6 Loetsch.Glacial Macun.Ground Loetsch.Ground Roseg.Ground 0. 6 1. 4 NMDS2 0 Roseg.Glacial Macun.Glacial 5 0.0 0.5 kryal krenal 0.0 0.5 NMDS1 1 -1.0 Loetsch.Glacial 0.4 -0.5 0.2 -0.5 NMDS2 70 65 55 1.2 NMDS2 5. Roseg.Glacial 80 1.4 NMDS2 4 Roseg.Glacial 90 Macun.Ground Loetsch.Ground Roseg.Ground 80 Roseg.Glacial 3 Macun.Glacial 60 20 2 10 0.5 40 30 0.5 kryal krenal Stress: 15 -1.0 -0.5 0.0 Stress: 15 0.5 -1.0 NMDS1 -0.5 0.0 NMDS1 44 0.5 Appendix Figure S7b 1 1 DOC [mg C L ] TIC [mg C L ] Sediment origin Sediment origin 6.7 % 14.4 % kryal krenal 3 0.5 0.5 kryal krenal 0. 5 2. 3.5 Macun.Glacial Macun.Glacial 4 5 1. 5 Macun.Ground Loetsch.Ground Roseg.Ground 5 0.0 NMDS2 Roseg.Glacial 0.0 NMDS2 1 5. 4.5 Roseg.Glacial Macun.Ground Loetsch.Ground Roseg.Ground 5 Loetsch.Glacial 2. -0.5 2 3 -0.5 5 3 Loetsch.Glacial Stress: 15 -1.0 -0.5 0.0 Stress: 15 0.5 -1.0 -0.5 0.0 NMDS1 0.5 NMDS1 1 1 NH4-N [ g L ] NO3-N [mg N L ] Sediment origin Sediment origin 27 % 10.7 % 0.5 kryal krenal 0.5 kryal krenal 22 20 Macun.Glacial Macun.Glacial 18 0.0 0.0 NMDS2 10 Loetsch.Glacial 0.16 8 Roseg.Glacial 0.16 Macun.Ground Loetsch.Ground Roseg.Ground 0. 15 Macun.Ground Loetsch.Ground Roseg.Ground 0.18 NMDS2 Roseg.Glacial 14 12 0.17 0.17 16 Loetsch.Glacial 0.15 4 0.14 6 0. 1 3 0. -0.5 -0.5 2 12 Stress: 15 -1.0 -0.5 0.0 Stress: 15 0.5 -1.0 NMDS1 1 Sediment origin 32.6 % 0.5 kryal krenal 2 2 0.0 Roseg.Glacial Macun.Ground Loetsch.Ground Roseg.Ground 1. 6 Loetsch.Glacial 1. 4 1. 2 1 -0.5 NMDS2 Macun.Glacial 1. 8 0. 8 Stress: 15 -1.0 -0.5 0.0 NMDS1 NO2-N [mg N L ] 2. -0.5 0.0 0.5 NMDS1 45 0.5 Appendix Figure S7c 860 1 1 DN [mg N L ] PN [mg N L ] Sediment origin Sediment origin 4.8 % 10.3 % 0.268 0.266 0.26Macun.Glacial 4 0.06 Macun.Glacial 0.252 Loetsch.Glacial 0.2 5 Roseg.Glacial Macun.Ground Loetsch.Ground Roseg.Ground Loetsch.Glacial 0.03 0.04 0.246 -0.5 -0.5 0.248 0.244 0.08 0.254 0.05 0.26 0.258 0.256 Macun.Ground Loetsch.Ground Roseg.Ground 0.0 NMDS2 Roseg.Glacial 0.0 NMDS2 0.262 0.07 0.5 kryal krenal 0.5 kryal krenal Stress: 15 -1.0 -0.5 0.0 Stress: 15 0.5 -1.0 -0.5 0.0 NMDS1 1 1 PO4-P [ g PL ] DP [ g PL ] Sediment origin Sediment origin 23.1 % kryal krenal 28 % kryal krenal 5. 5 6 5 4 7 6.5 0.5 5 0.5 0.5 NMDS1 Macun.Glacial 6 8 Macun.Glacial Roseg.Glacial 0.0 NMDS2 0.0 NMDS2 Macun.Ground Loetsch.Ground Roseg.Ground 5. 5 7 Loetsch.Glacial 4 3.5 2 2 -0.5 -0.5 Macun.Ground Loetsch.Ground Roseg.Ground 6 5 Loetsch.Glacial 4.5 2. 5 9 5 Roseg.Glacial 6 3 6. 3 Stress: 15 -1.0 -0.5 0.0 Stress: 15 0.5 -1.0 NMDS1 1 Sediment origin 22 % 0.5 kryal krenal Macun.Glacial Roseg.Glacial 0 0.0 25 Macun.Ground Loetsch.Ground Roseg.Ground 0 0 15Loetsch.Glacial 0 50 -0.5 NMDS2 300 100 Stress: 15 -1.0 -0.5 0.0 NMDS1 PP [ g PL ] 200 -0.5 0.0 0.5 NMDS1 46 0.5 Appendix Figure S8a 784500 785500 786500 787500 784500 785500 786500 787500 784500 785500 786500 DOC POC TIC 785500 786500 x coordinate (km) 787500 785500 786500 144000 142000 143000 VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A y coordinate (km) 144000 143000 784500 145000 x coordinate (km) 145000 x coordinate (km) 787500 784500 x coordinate (km) 785500 786500 143000 144000 145000 787500 784500 VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A 785500 786500 x coordinate (km) VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A x coordinate (km) 47 y coordinate (km) VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A x coordinate (km) VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A Organic matter 142000 142000 143000 VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A 144000 145000 D90D10 y coordinate (km) 145000 144000 143000 142000 y coordinate (km) VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A 142000 144000 143000 142000 y coordinate (km) 145000 784500 Conductivity y coordinate (km) 143000 144000 kryal krenal 142000 y coordinate (km) 145000 Temperature 787500 787500 Appendix Figure S8b 784500 785500 786500 787500 784500 785500 786500 787500 784500 785500 786500 143000 y coordinate (km) VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A 144000 145000 DN 142000 142000 143000 VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A 144000 145000 NO3.N y coordinate (km) 145000 144000 143000 142000 y coordinate (km) VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A 787500 784500 VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A 785500 786500 x coordinate (km) PN PO4.P DP PP 785500 786500 x coordinate (km) 787500 784500 785500 786500 787500 784500 x coordinate (km) 785500 786500 x coordinate (km) 865 48 144000 142000 143000 VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A y coordinate (km) 144000 142000 143000 VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A y coordinate (km) 144000 143000 VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A 145000 x coordinate (km) 145000 x coordinate (km) 145000 x coordinate (km) 142000 144000 143000 142000 y coordinate (km) 145000 784500 NO2.N y coordinate (km) 144000 143000 142000 y coordinate (km) 145000 NH4.N 787500 784500 787500 VR8A VR1A VR2A VR3A VR5A VR4A VR18A VR6A VR17A VR7A VR16A VR15A VR14A VR13A VR12A VR11A VR10A VR9A 785500 786500 x coordinate (km) 787500 Appendix Figure S8c 785500 786500 787500 784500 785500 786500 787500 784500 785500 786500 x coordinate (km) DOC POC TIC 785500 786500 x coordinate (km) 787500 784500 785500 786500 144000 142000 VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O 143000 VR8O VR1O VR2O y coordinate (km) 144000 VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O 143000 VR8O VR1O VR2O 145000 x coordinate (km) 145000 x coordinate (km) 787500 784500 x coordinate (km) 786500 145000 784500 VR8O VR1O VR2O VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O 785500 786500 x coordinate (km) VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O x coordinate (km) 49 787500 VR8O VR1O VR2O 785500 144000 VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O 143000 VR8O VR1O VR2O 142000 142000 VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O Organic matter y coordinate (km) y coordinate (km) VR8O VR1O VR2O 144000 145000 D90D10 143000 145000 144000 143000 142000 VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O 142000 144000 y coordinate (km) 143000 142000 784500 y coordinate (km) VR8O VR1O VR2O 145000 784500 Conductivity y coordinate (km) 143000 144000 kryal krenal 142000 y coordinate (km) 145000 Temperature 787500 787500 Appendix Figure S8d 785500 870 786500 787500 784500 785500 786500 787500 784500 785500 786500 145000 144000 VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O 143000 VR8O VR1O VR2O 142000 142000 VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O DN y coordinate (km) y coordinate (km) VR8O VR1O VR2O 144000 145000 NO3.N 143000 145000 144000 143000 142000 VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O 787500 784500 VR8O VR1O VR2O VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O 785500 786500 x coordinate (km) PN PO4.P DP PP 785500 786500 x coordinate (km) 787500 784500 785500 786500 787500 784500 x coordinate (km) 785500 786500 x coordinate (km) 50 144000 142000 VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O 143000 VR8O VR1O VR2O y coordinate (km) 144000 142000 VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O 143000 VR8O VR1O VR2O y coordinate (km) 144000 VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O 143000 VR8O VR1O VR2O 145000 x coordinate (km) 145000 x coordinate (km) 145000 x coordinate (km) 142000 144000 143000 142000 y coordinate (km) 784500 y coordinate (km) VR8O VR1O VR2O 145000 784500 NO2.N y coordinate (km) 144000 143000 142000 y coordinate (km) 145000 NH4.N 787500 784500 787500 VR8O VR1O VR2O VR5O VR4O VR18O VR6O VR7O VR16O VR15O VR14O VR13O VR12O VR11O VR10O VR9O 785500 786500 x coordinate (km) 787500 Appendix Figure S8e 784500 786500 787500 784500 786500 787500 784500 785500 786500 POC TIC 785500 786500 x coordinate (km) 787500 VR10J VR9J 785500 786500 144000 143000 142000 y coordinate (km) 144000 143000 784500 145000 DOC 145000 x coordinate (km) VR10J VR9J 787500 784500 x coordinate (km) 787500 VR10J VR9J 785500 786500 144000 143000 784500 VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J VR10J VR9J 785500 786500 x coordinate (km) VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J x coordinate (km) 51 145000 VR10J VR9J x coordinate (km) VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J y coordinate (km) VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J x coordinate (km) VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J Organic matter 142000 142000 VR10J VR9J 785500 143000 VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J 144000 145000 D90D10 y coordinate (km) 144000 145000 VR10J VR9J 785500 143000 142000 y coordinate (km) VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J 142000 144000 143000 142000 y coordinate (km) 145000 784500 Conductivity y coordinate (km) 143000 144000 kryal krenal 142000 y coordinate (km) 145000 Temperature 787500 787500 Appendix Figure S8f 784500 786500 787500 784500 786500 787500 784500 VR10J VR9J 785500 786500 143000 y coordinate (km) VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J 144000 145000 DN 142000 142000 VR10J VR9J 785500 143000 VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J 144000 145000 NO3.N y coordinate (km) 144000 145000 VR10J VR9J 785500 143000 142000 y coordinate (km) VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J 787500 784500 VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J VR10J VR9J 785500 786500 x coordinate (km) PN PO4.P DP PP 786500 x coordinate (km) 787500 786500 787500 x coordinate (km) VR10J VR9J 785500 786500 x coordinate (km) 52 144000 143000 y coordinate (km) 144000 784500 VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J 142000 VR10J VR9J 785500 143000 y coordinate (km) 144000 784500 VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J 142000 VR10J VR9J 785500 143000 VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J 145000 x coordinate (km) 145000 x coordinate (km) 145000 x coordinate (km) 142000 142000 143000 144000 145000 784500 NO2.N y coordinate (km) 144000 143000 142000 y coordinate (km) 145000 NH4.N y coordinate (km) 875 787500 784500 787500 VR8J VR1J VR2J VR3J VR5J VR4J VR18J VR6J VR17J VR7J VR16J VR15J VR14J VR13J VR12J VR10J VR9J 785500 786500 x coordinate (km) 787500 Appendix Figure S8g 633000 637000 633000 635000 POC TIC 635000 x coordinate (km) 637000 633000 635000 637000 x coordinate (km) 635000 x coordinate (km) 880 53 145000 y coordinate (km) L6A L5A L4A L2A L3A L1A 633000 L6A L5A L4A L2A L3A L1A 633000 635000 x coordinate (km) L7A L10A 143000 L6A L5A L4A L2A L3A L1A 637000 L9A L8A 144000 y coordinate (km) L7A L10A 143000 L6A L5A L4A L2A L3A L1A L9A L8A 145000 DOC 145000 x coordinate (km) L7A L10A 143000 143000 L6A L5A L4A L2A L3A L1A L9A L8A 144000 145000 144000 y coordinate (km) L7A L10A x coordinate (km) y coordinate (km) 144000 635000 Organic matter L9A L8A x coordinate (km) L7A L10A 143000 145000 637000 L9A L8A 633000 L6A L5A L4A L2A L3A L1A 144000 145000 635000 L7A L10A 143000 L6A L5A L4A L2A L3A L1A D90D10 L9A L8A 144000 y coordinate (km) L7A L10A 144000 145000 L9A L8A 633000 y coordinate (km) Conductivity kryal krenal 143000 y coordinate (km) Temperature 637000 637000 Appendix Figure S8h 633000 637000 633000 635000 145000 y coordinate (km) L7A L10A L6A L5A L4A L2A L3A L1A 143000 143000 L6A L5A L4A L2A L3A L1A L9A L8A 144000 145000 L7A L10A 144000 y coordinate (km) L9A L8A 637000 633000 635000 x coordinate (km) PN PO4.P DP PP 635000 x coordinate (km) 637000 633000 635000 637000 635000 x coordinate (km) 54 637000 637000 L9A L8A 144000 L7A L10A L6A L5A L4A L2A L3A L1A 143000 L6A L5A L4A L2A L3A L1A 633000 x coordinate (km) y coordinate (km) L7A L10A 143000 L6A L5A L4A L2A L3A L1A L9A L8A 144000 y coordinate (km) L7A L10A 143000 L6A L5A L4A L2A L3A L1A L9A L8A 145000 x coordinate (km) 145000 x coordinate (km) y coordinate (km) 144000 635000 DN x coordinate (km) L7A L10A 143000 145000 143000 637000 L9A L8A 633000 L6A L5A L4A L2A L3A L1A 145000 145000 635000 L7A L10A 144000 y coordinate (km) L6A L5A L4A L2A L3A L1A NO3.N L9A L8A 144000 145000 144000 L7A L10A 633000 y coordinate (km) NO2.N L9A L8A 143000 y coordinate (km) NH4.N 633000 635000 x coordinate (km) 637000 Appendix Figure S8i 635000 633000 635000 637000 633000 TIC 635000 x coordinate (km) 637000 633000 635000 637000 L6O L5O L4O L1O 633000 x coordinate (km) 635000 x coordinate (km) 55 145000 L6O L5O L4O L1O 633000 635000 x coordinate (km) L7O L10O 143000 L1O 637000 L9O 144000 y coordinate (km) L6O L5O L4O 143000 L1O 144000 y coordinate (km) L6O L5O L4O 145000 POC 145000 DOC 145000 x coordinate (km) L7O L10O L7O L10O 143000 635000 x coordinate (km) L9O y coordinate (km) L1O L9O 144000 145000 L6O L5O L4O 143000 L1O L7O L10O 144000 y coordinate (km) L6O L5O L4O Organic matter L9O x coordinate (km) L7O L10O 144000 145000 637000 L9O 633000 L7O L10O 143000 L1O D90D10 L9O 144000 L6O L5O L4O 143000 y coordinate (km) y coordinate (km) L7O L10O 144000 145000 L9O 633000 885 Conductivity kryal krenal 143000 y coordinate (km) Temperature 637000 637000 Appendix Figure S8j 635000 637000 637000 633000 145000 L7O L10O L6O L5O L4O L1O 143000 L1O 143000 635000 y coordinate (km) L6O L5O L4O L9O 144000 145000 L7O L10O 144000 y coordinate (km) L9O 635000 637000 633000 635000 PN PO4.P DP PP 635000 x coordinate (km) 637000 633000 635000 637000 635000 x coordinate (km) 56 L7O L10O L6O L5O L4O L1O 143000 L1O 637000 637000 L9O 144000 L6O L5O L4O 633000 x coordinate (km) y coordinate (km) L7O L10O 143000 L1O L9O 144000 y coordinate (km) L6O L5O L4O 143000 L1O L7O L10O 144000 y coordinate (km) L6O L5O L4O L9O 145000 x coordinate (km) 145000 x coordinate (km) 145000 x coordinate (km) 145000 144000 L1O DN x coordinate (km) L7O L10O 143000 L6O L5O L4O 633000 L9O 633000 L7O L10O 143000 L1O 145000 y coordinate (km) L6O L5O L4O NO3.N L9O 144000 145000 144000 L7O L10O 633000 y coordinate (km) NO2.N L9O 143000 y coordinate (km) NH4.N 633000 635000 x coordinate (km) 637000 Appendix Figure S8k 635000 633000 637000 633000 635000 POC TIC 635000 x coordinate (km) 637000 633000 635000 637000 x coordinate (km) 635000 x coordinate (km) 57 145000 y coordinate (km) L6J L5J L4J L2J L3J L1J 633000 L6J L5J L4J L2J L3J L1J 633000 635000 x coordinate (km) L7J L10J 143000 L6J L5J L4J L2J L3J L1J 637000 L9J L8J 144000 y coordinate (km) L7J L10J 143000 L6J L5J L4J L2J L3J L1J L9J L8J 145000 DOC 145000 x coordinate (km) L7J L10J 143000 143000 L6J L5J L4J L2J L3J L1J L9J L8J 144000 145000 144000 L7J L10J x coordinate (km) y coordinate (km) 144000 635000 Organic matter L9J L8J x coordinate (km) L7J L10J 143000 145000 637000 L9J L8J 633000 L6J L5J L4J L2J L3J L1J 144000 145000 633000 L7J L10J 143000 L6J L5J L4J L2J L3J L1J y coordinate (km) L7J L10J D90D10 L9J L8J 144000 y coordinate (km) L9J L8J 144000 145000 Conductivity kryal krenal 143000 y coordinate (km) Temperature y coordinate (km) 890 637000 637000 Appendix Figure S8l 633000 637000 633000 635000 145000 y coordinate (km) L7J L10J L6J L5J L4J L2J L3J L1J 143000 143000 L6J L5J L4J L2J L3J L1J L9J L8J 144000 145000 L7J L10J 144000 y coordinate (km) L9J L8J 637000 633000 635000 x coordinate (km) PN PO4.P DP PP 635000 x coordinate (km) 637000 633000 635000 637000 635000 x coordinate (km) 895 58 637000 637000 L9J L8J 144000 L7J L10J L6J L5J L4J L2J L3J L1J 143000 L6J L5J L4J L2J L3J L1J 633000 x coordinate (km) y coordinate (km) L7J L10J 143000 L6J L5J L4J L2J L3J L1J L9J L8J 144000 y coordinate (km) L7J L10J 143000 L6J L5J L4J L2J L3J L1J L9J L8J 145000 x coordinate (km) 145000 x coordinate (km) y coordinate (km) 144000 635000 DN x coordinate (km) L7J L10J 143000 145000 143000 637000 L9J L8J 633000 L6J L5J L4J L2J L3J L1J 145000 145000 635000 L7J L10J 144000 y coordinate (km) L6J L5J L4J L2J L3J L1J NO3.N L9J L8J 144000 145000 144000 L7J L10J 633000 y coordinate (km) NO2.N L9J L8J 143000 y coordinate (km) NH4.N 633000 635000 x coordinate (km) 637000 Appendix Figure S8m 806000 806500 805500 806000 179000 805000 805500 806000 805000 805500 806000 x coordinate (km) 806500 M16A M15A 805000 805500 806000 806500 178800 M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A 178600 178200 M10A M13A M12A M14A M11A M17A 178400 y coordinate (km) 178800 178600 178200 178400 y coordinate (km) 178800 M16A M15A M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A M10A M13A M12A M14A M11A M17A M16A M15A 805000 x coordinate (km) 805500 806000 x coordinate (km) 59 178800 M10A M13A M12A M14A M11A M17A M16A M15A 805000 805500 806000 x coordinate (km) 179000 TIC 179000 POC 179000 DOC 806500 M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A 178600 178200 M16A M15A x coordinate (km) M10A M13A M12A M14A M11A M17A y coordinate (km) 178800 178600 806500 178400 179000 805000 178400 y coordinate (km) M16A M15A M10A M13A M12A M14A M11A M17A x coordinate (km) M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A Organic matter M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A x coordinate (km) 178600 178400 178200 M10A M13A M12A M14A M11A M17A 178200 M16A M15A 805500 178800 178200 M10A M13A M12A M14A M11A M17A M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A 178600 y coordinate (km) 178800 M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A 805000 y coordinate (km) D90D10 179000 Conductivity 178400 kryal krenal 178600 178400 178200 y coordinate (km) 179000 Temperature 806500 806500 Appendix Figure S8n 805500 806000 805000 805500 806000 806500 179000 M16A M15A 805000 805500 178800 178200 M10A M13A M12A M14A M11A M17A 806000 M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A 178600 y coordinate (km) 178800 178600 178400 y coordinate (km) M16A M15A M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A 178400 179000 DN 806500 M10A M13A M12A M14A M11A M17A M16A M15A 805000 805500 806000 PN PO4.P DP PP 805000 805500 806000 x coordinate (km) 806500 805500 806000 806500 M16A M15A 805500 806000 x coordinate (km) 60 806500 178800 M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A 178600 178400 M10A M13A M12A M14A M11A M17A 805000 x coordinate (km) y coordinate (km) 178600 178800 M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A 178200 M16A M15A 805000 178400 y coordinate (km) 178800 M10A M13A M12A M14A M11A M17A 178200 M16A M15A M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A 178600 178200 M10A M13A M12A M14A M11A M17A 178400 y coordinate (km) 178800 M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A 806500 179000 x coordinate (km) 179000 x coordinate (km) 179000 x coordinate (km) 179000 x coordinate (km) 178600 178400 178200 y coordinate (km) 178800 806500 M10A M13A M12A M14A M11A M17A 178200 M16A M15A M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A 178600 178200 M10A M13A M12A M14A M11A M17A 178400 y coordinate (km) 178800 178600 178400 178200 y coordinate (km) M9A M5A M7A M6A M8A M1A M3AM4A M2O M2A 805000 900 NO3.N 179000 NO2.N 179000 NH4.N M10A M13A M12A M14A M11A M17A M16A M15A 805000 805500 806000 x coordinate (km) 806500 Appendix Figure S8o 805500 806000 806500 805500 806000 806500 179000 805000 805500 806000 TIC M10O M12O M14O 805500 806000 x coordinate (km) 806500 178800 178600 M3OM4O M10O M12O M14O 178200 178200 805000 805000 805500 806000 806500 805000 x coordinate (km) 805500 806000 x coordinate (km) 61 178800 178600 M10O M12O M14O 805000 805500 806000 x coordinate (km) M9O M7O M6O M8O 178400 y coordinate (km) 178800 178600 M3OM4O 178400 y coordinate (km) 178800 178600 178200 178400 M10O M12O M14O 806500 179000 POC 179000 DOC 179000 x coordinate (km) M9O M7O M6O M8O M3OM4O 178200 178200 805000 x coordinate (km) M3OM4O y coordinate (km) 178800 178600 M10O M12O M14O M9O M7O M6O M8O 178400 179000 M10O M12O M14O M3OM4O x coordinate (km) M9O M7O M6O M8O Organic matter M9O M7O M6O M8O 178400 178800 M3OM4O 178200 178400 M10O M12O M14O y coordinate (km) 178600 M3OM4O M9O M7O M6O M8O 178600 y coordinate (km) 178800 M9O M7O M6O M8O 178400 kryal krenal 805000 y coordinate (km) D90D10 179000 Conductivity 178200 y coordinate (km) 179000 Temperature 806500 806500 Appendix Figure S8p 805500 806000 806500 179000 M10O M12O M14O 805500 806000 806500 178800 M10O M12O M14O 178200 178200 805000 M3OM4O 178600 y coordinate (km) 178800 178600 y coordinate (km) M3OM4O M9O M7O M6O M8O 178400 179000 DN M9O M7O M6O M8O 178400 178800 M10O M12O M14O 178200 805000 805000 805500 806000 806500 805000 805500 806000 PN PO4.P DP PP 805500 806000 x coordinate (km) 806500 805000 805500 806000 806500 805000 x coordinate (km) 805500 806000 x coordinate (km) 62 178800 178600 M3OM4O M10O M12O M14O 178400 178800 M10O M12O M14O M9O M7O M6O M8O 178200 178200 805000 y coordinate (km) M10O M12O M14O M3OM4O 178600 178800 y coordinate (km) M10O M12O M14O M3OM4O M9O M7O M6O M8O 178200 178600 M3OM4O M9O M7O M6O M8O 178600 y coordinate (km) 178800 M9O M7O M6O M8O 806500 179000 x coordinate (km) 179000 x coordinate (km) 179000 x coordinate (km) 179000 x coordinate (km) 178400 y coordinate (km) 178600 y coordinate (km) 178400 M10O M12O M14O M3OM4O 178400 920 NO3.N M9O M7O M6O M8O 178400 179000 178800 178600 M3OM4O 178200 y coordinate (km) M9O M7O M6O M8O 178200 915 NO2.N 179000 NH4.N 910 178400 905 806500 805000 805500 806000 x coordinate (km) 806500 Figure S9a 925 0.2 Env PCoA M14A M17A M8A M10A M9A M7O M7A M3A M6A M3O M6O M8O M4A M11A M14O M12A M2A M10O M4O M9O M2O M5A M16A M15A VR13O VR10A VR12A VR14A VR13A VR9A VR11A -0.1 0.0 VR17A M12O VR3A L7J L7O VR2A VR15A L7A VR7A VR18A L8J L10A L9A VR5A VR12O VR18J L10J L9J L6A L9O VR18O VR8A L10O L4A VR10O L8A L6O L1J L3J L2J L4O L6J L4JVR9O L5A VR15J L5JL3A VR12J VR16A VR9J VR1A VR6A VR10J VR16J VR5O VR16O VR8O VR7O VR5J L2A L1A L5OVR1O VR14J VR2J VR13J VR15O VR17J VR3J VR1J VR7J VR8J VR2O VR6J VR11O VR14O VR4O VR6O VR4A L1O VR4J -0.2 PCoA2 0.1 M1A M13A -0.1 0.0 0.1 PCoA1 63 0.2 Figure S9b 0.3 0.4 Enz PCoA M10A M6A VR16A M6O M2O M14A 0.2 VR1A 0.1 VR2A L6A VR14JL6J M8O M2A M7O VR8O M14O M8A M4A M16A M1A M3A VR1O VR7JL4J M5A M15AM4O M10O L6O VR11A M13A M12O VR12A VR16J VR1J M12A VR15O VR5O VR16O VR11O VR13A M17A VR4A M9O M11A M3O VR14AL4O VR3A VR4O L4A VR9A VR5A VR8A VR7A VR10A L5O VR10J L3A VR10O VR9J L10A L7A L5A VR4J VR15J L9O VR18OVR6A L5J VR5J VR7O L10O VR15A L7O VR14O VR9OL1A L1J L10J L7J L3J L2J VR2O L9A VR12OVR8J VR17A VR2J VR6J L8J VR13O VR18J L2A L8A VR18AL9J VR3J VR17J L1O VR6O -0.2 -0.1 0.0 VR12J VR13J -0.3 PCoA2 M9A M7A -0.4 -0.2 PCoA1 64 0.0 0.2 930 Figure S9c 0.4 Com PCoA L10O L10J L9JL9A L10A 0.3 L9O 0.2 VR3J VR3A L2A 0.1 0.0 -0.1 -0.2 PCoA2 VR15A L2J VR15O VR15J VR2J VR17J VR6J L5J VR18A L7A VR4J L8J L5A M4A L7OL8A M8O VR18O VR6A VR16A VR18J VR6OM7O VR16O VR16J M3OM4O L7J VR1A VR1O VR4A L3J M8A M13A VR1J VR4O M3A M11A M9O M6O M15A M12O M9A M2A M12A M2OM17AM16A M7AM6A M10A M14O M14A M10O M1A VR5A M5A L4J VR9J VR2A L6O L5O VR10J VR8O VR8J L1J VR17A VR8A VR5J L1O VR10O L4O VR5O L4A VR2O VR13J VR13A VR13O VR7O VR9A VR7J VR9O VR14J L3A L1A VR14O VR14A VR12O VR7A VR11O VR12J L6A VR10A L6J VR12A VR11A -0.2 0.2 0.0 PCoA1 65 0.4