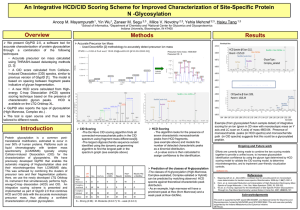
pmic7061-sup-0001-FigureS1
advertisement

1 Supplemental Information 2 3 Materials and Methods 4 Standard laboratory reagents were purchased from Sigma, Roth and Merck unless otherwise 5 noted. The commercial bovine serum albumin digest was purchased from Michrom 6 Bioresources. 7 The E. coli digest was generated by hypotonic cell lysis followed by isolating the cytosolic 8 fraction by centrifugation and subsequent digestion with trypsin at an enzyme to substrate 9 ratio of 1:100. 10 For the lapatinib selectivity assay, proteins were extracted from human placenta and kinases 11 were purified in the presence and absence of 1 µM lapatinb (LC laboratories, MA) as 12 described (Bantscheff et al Nat. Biotechnol, 2007). 13 TMT labeling was performed as described (Bantscheff et al Nat. Biotechnol, 2007). 30 ng of 14 TMT labeled BSA digest (TMT 126:127:128:129:130:131 = 1:2:4:8:16:32) was spiked into 6 15 μg TMT-labeled E.coli digest (1:1:1:1:1:1). 16 17 HPLC and Mass Spectrometry 18 Mass spectrometry was performed on a LTQ Orbitrap XL mass spectrometer (Thermo Fisher 19 Scientific, Germany) coupled to a nanoLC Ultra 1D+ liquid chromatography system 20 (Eksigent, CA). Peptides were separated using an in house packed trap column (20 mm x 75 21 μm RepoSil-Pur C18, Dr. Maisch, Germany) in line with an analytical column (200 mm x 75 22 μm RepoSil-Pur C18, Dr. Maisch, Germany) and gradient elution was performed from 2 to 23 28% solvent B (0.1 M formic acid in 100% acetonitrile) within 4 h at a flowrate of 300 nl/min. 24 The eluent was sprayed via emitter tips (PicoTip, New Objective, MA) using a nano- 25 electrospray ion source (Proxeon Biosystems, DK). The mass spectrometer was operated in 1 1 positive ion mode. Full scan MS spectra were acquired in the orbitrap recording a window 2 between 300 and 1500 m/z at a resolution of 60,000 (at m/z 400) after accumulation to a 3 target value of 500,000. CID of the five most intense ions was performed in the LTQ at a 4 normalized collision energy of 35% after accumulation to a target value of 10,000 for max 5 500 ms. Subsequently, HCD tandem mass spectra were triggered from the same five 6 precursor ions. HCD ions were generated in the HCD collision cell using a normalized 7 collision energy of 75%, a target value of 30,000 and 750 ms max accumulation time and 8 subsequently detected in the orbitrap at 7500 resolution. Precursor ions were put on a 9 dynamic exclusion list for 30 s. Internal calibration was enabled for both MS and MS/MS 10 mode using the ion signal of (Si(CH3)2O)6 as a lock mass. 11 12 Data processing 13 The raw CID-HCD data was processed into Mascot searchable files (mgf) using Mascot 14 Distiller (Matrix Science, UK). Two separate .mgf files were generated, each with peak 15 processing and picking optimized for either peptide identification by CID or TMT 16 quantification by HCD spectra. Briefly, uncentroiding of tandem MS spectra and isotope 17 fitting was enabled for optimal CID scan processing (_CID.mgf). For HCD scans, peak lists 18 were generated without further peak processing (_HCD.mgf). Then, the two .mgf files were 19 merged into a single .mgf file using the provided Perl script (CID-HCDmerge). In short, for 20 each HCD spectrum the TMT reporter ions were detected by their characteristic mass 21 differences. The intensities of the reporter ions were extracted and their mass offset 22 corrected. Further processing and visualization of the TMT intensities was done in MATLAB 23 (The MathWorks, Germany). The processed reporter signals were pasted into the 24 corresponding CID spectrum of the _CID.mgf file, deleting the respective m/z window at the 25 same time (for more detail please see supplemental figure S1). Resulting peak lists 26 (_CID_cut_HCD.mgf) were searched using the Mascot search engine version 2.3.01 (Matrix 2 1 Science, UK). Data from the BSA/E.coli experiment was searched against the SwissProt 2 database version 57.15 (515,203 entries). Data from the human samples were searched 3 against the human International Protein Index (IPI) database version 3.78 (87,061 entries). A 4 precursor tolerance of 10 ppm and a fragment tolerance of 0.6 Da was used in all cases. 5 Enzyme specificity was set to trypsin and up to two missed cleavage sites were allowed. 6 TMT 6plex at N-termini and lysine residues was set as fixed modification and, for the 7 lapatinib selectivity data set, also carbamidomethylation at cystein. Variable modifications 8 included N-terminal acetylation, oxidation of methionine, phosphorylation of serine, threonine 9 and tyrosine and pyroglutamic acid formation of glutamine and glutamic acid, for the 10 BSA/E.coli experiment in addition carboxy- (BSA) and carbamido- (E.coli) methylation at 11 cystein. Quantification of TMT-labeled peptides was performed by Mascot without any further 12 isotope corrections or normalization (already done within the pearl script). 13 14 Other notes 15 It should be noted that the pearl script we provide will only work for Mascot mgf files 16 generated by Mascot Distiller from LTQ Orbitrap XL raw data files using a top1-5 CID 17 followed by top1-5 HCD of the same precursors. Any other acquisition schemes and mgf 18 processing will require modifications to the script (source code is provided). We provide the 19 source code (file name: CID_HCD_Merge) and version compiled for Windows (file name: 20 Win32_CID_HCD_Merge, which also contains a readme file describing how to run the 21 script). 22 23 3 1 Supplemental figures 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 Figure S1: Configuration file of the provided script for processing data from combined CIDHCD acquisitions. Input data for the script are two .mgf files (_CID.mgf and _HCD.mgf), both processed by Mascot distiller for the respective scan types. TMT6plex-labeled raw data must be recorded in combined CID-HCD mode where up to five CID scans are followed by the HCD scans from the same precursor. First the two corresponding spectra are identified by their precursor mass within a window of 10 scans. TMT reporter ions are then detected in the _HCD.mgf by their characteristic mass differences (theoretical mass differences calculated from configured masses +/- tolerance distol) and their m/z values are corrected by the median offset of all TMT signals within the spectrum. All identified peaks are extracted, their intensities adjusted by the hcdfactor parameter and submitted to the Matlab software (integrated in the merge script) for further peak processing to improve quantification. This processing comprises isotope impurity correction of the isobaric tags by factors defined in the configuration file, discarding of scans with missing reporter channels (tolmissvalfromleft; most important for missing reference signals) and different normalization methods. Moreover signal intensity distributions within their m/z value and against reference intensities are visualized to estimate data quality. In a final step the refined reporter signals are merged in the respective CID spectrum in the _CID.mgf where all peaks in a certain mass range around the extracted ions (cidcuttol) are discarded. The resulting file format is again a .mgf file with ‘chimeric’ CID-HCD spectra for identification and quantification via Mascot. Further detail: the top paragraphs relate to which fragment ions will be extracted from a given HCD spectrum. A total of 9 TMT related ions are listed which include the six actual TMT channels as well as a three related ions (125 for incomplete stable isotope labeling of the TMT126 reagent; 132 and 133 for the natural C13 isotopes of the TMT 131 reagent). In addition to the TMT ions, any other ion of interest may be extracted from the HCD spectrum. Some immonium ions of amino acids and the oxonium ion of HexNAc are shown as examples. The next section lists the actual processing parameters (see above) which related to the mass tolerances with which the difference processing and the HCD extraction and CID insertion are performed. The script allows restricting processing up to a specified maximum LC retention time. The remaining parameters relate to TMT quantification. Because the isotopic impurities of all TMT 4 1 2 3 4 reagents, the relative contributions of one TMT channel to the others can be corrected. The correction values are provided by the vendor and the script will optionally perform this step. It is further possible to account for missing TMT channels and to normalize the data by two different methods. 5 5 1 2 3 4 5 6 Figure S2: TMT mass region extracted from data collected by LC.-MS/MS on a qTOF instrument (ABSciex tripleTOF, see also figure 2 of the main manuscript). The data shows that several additional ion signals are present in the TMT region but the presence of interfering signals is a lot less pronounced than for the LTQ-Orbitrap XL. TMT ions are detected with mass errors between 8 and 15 ppm. 7 6 1 2 3 Figure S3: similar data as shown in figure 2A but when using two different batches of the TMT reagents. 4 7 1 2 3 4 5 6 7 8 Figure S4: Quantification distortion by the presence of near-isobaric signals. The example spectrum (unprocessed data) shows the presence of one non-TMT ion near the TMT-128 channel and two such ions near the TMT-131 channel. When processing this data by Mascot Distiller (our standard for processing raw Orbitrap files into Mascot searchable mgf files), The TMT-128 intensity is erroneously increased. For the TMT-131 channel, both the processed m/z value and the intensity are distorted. Mass difference processing of the raw data avoids this distortion. 9 8 1 2 3 Table S1: Possible chemical compositions calculated for the m/z species detected in the TMT reporter ion region of the HCD data shown in figure 2. 4 5 6 9