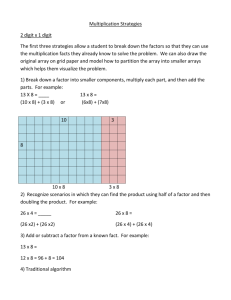
Number of samples
Microarray Sample Submission Form
Project title (with one sentence):
____________________________________________________________________
Project number (filled by FMSC):
Project description: ____________________________________________________
____________________________________________________________________
____________________________________________________________________
____________________________________________________________________
Contact information
Contact person(s)
Name(s):
e-mail(s):
Address:
Phone no.:
BioCity Turku Reasearch Programme (filled by Biocity Turku groups):
__________________________________________________________________
Group leader / Principal investigator
Name:
Address:
Phone no.:
e-mail:
Invoice information:
Internal invoice (University of Turku account)
Name:
Institution:
Department/laboratory:
Unit code:
External invoice (other than University of Turku account)
Institution:
Department/laboratory:
Address:
Your VAT number:
Project Opening Meeting
has been held on ____________________ (date )
Participants :__________________________________________________________
______________________________________________________________________
______________________________________________________________________
Array platform:
Illumina arrays, please specify array type: _________________________________
Agilent arrays, please specify array type: __________________________________
Affymetrix arrays, please specify array type: _______________________________
You can find more information about the array types in our web pages
( http://fmsc.btk.fi
). If you have any questions about array content, please contact us.
Sample information:
Number of samples: ____________
Organism: ____________________________________________________
Tissue,origin: ____________________________________________________
Cells, origin: ____________________________________________________
Blood, origin: ____________________________________________________
Other,what: ____________________________________________________
RNA:
totalRNA
mRNA
RNA extraction method:
______________________________________________________________________
______________________________________________________________________
DNase treatment method (we expect that all delivered RNA samples are Dnase-treated.
In other case contact us to discuss this):
______________________________________________________________________
______________________________________________________________________
RNA is dissolved in:
NOTE:
COMPARISON TO A SET OF PREVIOUS RESULTS
If you are planning to c ompare these array results with a previous set of samples done using our service, please indicate here the time when the previous samples were processed:
COMPARISON TO A SET OF FUTURE RESULTS
If you are planning to compare these array results directly with a possible future set of samples done using our service and the same array type, please indicate this here and also give us some estimate when these samples will be processed.
(It should be noted that after longer time periods all array types and protocols may not be available anymore, complicating the integration between earlier and later data set) :
LEFT OVER MATERIAL DISPOSAL
FMSC will discard both the original samples and their labeling products after 1 year. We prefer receiving only the amount of sample needed for the analysis but please indicate here if you for some reason need to send extra material and wish to have it returned. In such case please note that the customer has to cover the possible costs related to the returning of the sample material.
STORING THE RESULT DATA:
Analysis results will be sent to the customer electronically and after this the customer is responsible for storing the result data . FMSC will store especially the large raw image data files only for a short time. As the extracted numerical raw data files are delivered for the customer, the original image files rarely have further use. In case you would also like to receive the raw image data files, please this indicate here and we will send them to you on
CDs/DVDs:
Sample Information:
Use less than 32 characters. Avoid spaces and any special characters in the sample names. Please use group names and sample names that will allow an easy interpretation of the name coding during data analysis. (Add rows if needed.)
Sample Sample name
Sample
Group c, ng/
l A
260
/A
280
Volume,
l Comments
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
Group desciptions
Please give a short description of each sample group (add rows if needed)
Sample Group Num. of replicates
Description
Changing your sample setup:
Please note that if you have changed your original sample setup from the one discussed with our bioinformaticians, you should contact them ( bioinfo@btk.fi) to discuss the changes before continuing further. This concerns especially those projects where the FMCS bioinformatics data analysis service is used.
2
3
4
5
Data analysis service
____ I would like to purchase the full bioinformatics data analysis service
____ The data will be analyzed through a collaboration with the FMSC bioinformatics group (as agreed previously)
____ I will analyze the data myself, data will be sent to this e-mail address:
___________________________________
In case the FMSC's bioinformatics team will analyze the data fill in:
Direct comparisons between groups:
(Remember that all groups do not necessarily need to be directly compared but the lists of differentially expressed genes between several independent comparisons (e.g. for different tissues) can also be compared.)
Use group names given in the sample information table.
Comparison Group1 Group2
0 (example) WT (wild type group)
1
KO (knock-out group)
6
7
Paired samples:
Please indicate clearly (by e.g. sample naming) the samples that originate from the same biological individual as this information is needed in the analysis.
Other information for the bioinformatic analysis (e.g. other factors linking certain samples together like handling day etc.):
Please, send us the form both in print with your samples and electronically as an e-mail attachment . We require a signed copy of this document prior to processing the samples.
As Principal Investigator on this project, I agree to:
1.
Acknowledge the use of the Finnish Microarray and Sequencing Centre's analysis service in the materials and methods section in any resulting publications.
2.
Send a reprint of each resulting publication to the the Finnish Microarray and
Sequencing Centre.
3.
Include the FMSC's bioinformatics team in my resulting publication(s) (where the analysis results are used) as coauthors if a collaboration has been agreed
Date with them. I will also send the manuscript for their review prior to submitting to a journal.
Signature