Supplemental Methods
advertisement

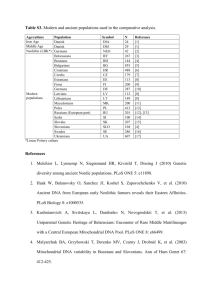
AFFYMETRIX MICROARRAY All adipose tissues used in the study were inguinal subcutaneous adipose tissue. At sacrifice, the adipose tissue (100 to 350 mg) was harvested, minced finely, placed into 5~10 volumes of RNALater™ (Ambion, Inc., Austin, TX), and stored in a -80C freezer until further processing. Tissue from RNALater™ stocks was weighed, transferred to Trizol reagent (Invitrogen, Carlsbad, CA), and homogenized using the Mixer Mill system (Retsch, Haan, Germany). The clear aqueous supernatant was transferred to RNeasy Mini columns (Qiagen Inc., Valencia, CA) and processed (including DNase I treatment) according to manufacturer’s instructions. RNA integrity was assessed by OD260/280 ratio and 28S and 18S ribosomal RNA ratio with Agilent BioAnalyzer (Agilent Technologies Inc., Palo Alto, CA). All RNA samples were randomized to avoid batch effects. Total RNA (5 g) from single animals was individually converted into biotinylated cRNA using protocols recommended by the microarray manufacturer (Affymetrix). Labelling quality was assessed by cRNA yields and integrity monitored by Agilent BioAnalyzer. The cRNA samples were re-randomized. Hybridization cocktails containing 10 g of fragmented cRNA from single animals were loaded onto GeneChip® Mouse Genome 430A Array and hybridized overnight. Genechips® were washed and scanned using Affymetrix fluidic stations and scanners. Intensity data were captured by Genechip Computer Operating System using the algorithm, MAS 5.0 (Affymetrix Microarray Suite, Santa Clara, CA), and probe sets with an intensity > 22.1 were considered present in the tissue. IMMUNOHISTOCHEMISTRY Inguinal adipose tissue fixed in 10% neutral buffered formalin was paraffin-embedded and sectioned (5-m in thickness). The sections were deparaffinized and microwaved (10 min) in Target Retrieval Solution pH6 (DAKO, Glostrup, Denmark) for antigen retrieval as previously described (1). The sections were then quenched with Peroxidase Blocking Solution (DAKO; 10 min), and blocked with 1% normal goat or rabbit serum (20 min). Tissue sections were incubated (60 min) with goat anti-cytochrome c (1:500) (Santa Cruz, Santa Cruz, CA), rabbit anti-UCP1 (1:2000) (Abcam, Cambridge, UK), or goat anti-Hsp60 polyclonal antibody (1:200) (Santa Cruz, Santa Cruz, CA). After washing with Wash Buffer, the sections were incubated with biotinylated rabbit anti-goat or goat anti-rabbit IgG antibody (Vector Laboratories; 30 min), and then ABC reagent of VECTASTAIN (Vector laboratories; 30 min). The reaction product was visualized by incubation with DAKO Liquid DAB Substrate Chromogen (2 min) and immersed in distilled water (3 min). The sections were counterstained with hematoxylin (2 min), rinsed with distilled water, dehydrated, mounted with Entellan New (MERCK) and cover-slipped. MORPHOMETRIC ANALYSIS Quantification of cytochrome c-positive areas was performed using eCognition (Definiens AG, Germany). Five images from one section (total 0.705 mm2 area per slide; 5 sections per animal) were randomly acquired at x200 magnification using a light microscope with DP70 system (Olympus, Tokyo, Japan) by two independent, blinded pathologists. The acquired images were segmented using the algorithm of chessboard segmentation into equal-sized (1 pixel) squares according to manufacturer’s instruction (User Guide Cellenger Developer Studio 4.0, 2004; www.definiens.com). The segmented objects with a blue image layer value < 160 were temporary selected as cytochrome cpositive area, and then the objects with a red image layer value < 150 and blue image layer value > 100 were excluded from the final classification. Cytochrome c-positive area per section was calculated as cytochrome c-positive area divided by the total area of examined images, and expressed as m2/mm2. UCP1-positive multilocular cells on acquired images were identified and counted by blinded pathologists, and expressed as number of cells per 0.705 mm2. QUANTIFICATION OF RNA AND MITOCHONDRIAL DNA BY QUATITATIVE REAL TIME PCR Adipose RNA was reverse-transcribed using ABI High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA) to make cDNA. Total adipose DNA was extracted from homogenized inguinal adipose tissue (without RNALater™) according to the manufacturer’s protocol (Promega, Madison, WI). The cDNA and DNA were subject to quantitative real-time PCR (10 and 100 ng per reaction for cDNA and DNA, respectively) as previously described (2), carried out with Universal PCR Master Mix 2X (Applied Biosystems) using 7900HT Sequence Detector System (Applied Biosystems) as recommended by the vendor. The primer/probe sequences are listed in the following table, except Cidea (cell death-inducing DNA fragmentation factor, alpha subunit-like effector A), which was ordered directly from Applied Biosystems. The relative abundance of a target gene RNA was normalized to both housekeeping genes beta actin (Actb) and 36B4 (Arbp), and expressed as fold changes as indicated in Tables 2A and 2B in the text. Relative amounts of nuclear and mitochondrial DNA were determined by Gapdh and cytochrome b, respectively, and the ratio of mitochondrial DNA to nuclear DNA reflects the tissue concentration of mitochondria per cell (2). Symbols Gene Name Forward Primer Reverse Primer PROBE Acadm acetyl-Coenzyme A dehydrogenase, medium chain TGACGGAGCAGCCAATGA ATGGCCGCCACATCAGA TGCTTACTGTGTGACAGAGCCCTCCG Actb beta actin GAGCTATGAGCTGCCTGACG AGTTTCATGGATGCCACAGGA CATCACTATTGGCAACGAGCGGTTCC Arbp 36B4 (acidic ribosomal phosphoprotein P0) GCTTCATTGTGGGAGCAGACA TGCGCATCATGGTGTTCTTG TCCAAGCAGATGCAGCAGATCCGC Cat catalase GGATTATGGCCTCCGAGATCT TAAAACGTCCAGGACGGGTAA TTCAATGCCATCGCCAATGGCA Cpt1b carnitine palmitoyltransferase 1b, muscle AGGGCTCGCGCCTTCT GAAGGGTCGTCGAGGATTCTC CCCGAGACCTGGAGATGCAGTTC Cs citrate synthase CCCCTGCCTGAGGGCTTAT GCCAAGACACCTGTTCCTCTGT TGGCTGCTGGTAACTGGACAGATGCC Cyc1 cytochrome c-1 CCTGCCACAGCATGGATTATG TTGGCTTCTTCCTCCGTGTAG CGTACCGCCACCTGGTGGGAGT Cycs cytochrome c, somatic GCAAGCATAAGACTGGACCAAATC ATGCCTTTGTTCTTGTTGGCATC TAAGAGAATCCAGCAGCCTGGCCTGTC Dld dihydrolipoamide dehydrogenase TTGATACCAAAAACATCCTTGTAGCT AGTATCTTCATCGATCGTGATTCCA CGGGCTCAGAAGTTACTCCTTTTC Hspa9a mitochondrial heat shock protein 70 GCAAAGGTCCTGGAGAATGCT TCGTTCTCCATCTGCTGTAAAGG AAGGTGCCAGAACTACCCCTTCTGTGGT Hspd1 mitochondrial heat shock protein 60 CCGAAGACGTTGACGGAGAA TGCCACAACCTGAAGACCAA CTCTAAGCACGCTGGTTTTGAACAGGCTAAA Immt inner membrane protein, mitochondrial CTGCCTGCCGTCGAAGTT AGATGCCATGGAGAATGAAATGCGGAC Mrpl15 mitochondrial ribosomal protein L15 AAGCCCAGTCCTAACTCCAGAA TCTGCCACACTTCCTACCTCTTC ACGGGAAAGACGTCCAAGAGATCGG Opa1 optic atrophy 1 homolog TTGCCTGGGAGACTCTACAAGAG AATATGTCGTCGTGTTCCTTTCC TTTCCCGCTTCATGACAGAACCCAA Ppargc1a peroxisome proliferative activated receptor, gamma, coactivator 1 alpha CATTTGATGCACTGACAGATGGA CCGTCAGGCATGGAGGAA CCGTGACCACTGACAACGAGGCC Ppargc1b peroxisome proliferative activated receptor, gamma, coactivator 1 beta AGGAAGCGGCGGGAAA CTACAATCTCACCGAACACCTCAA AGAGATTTCGAATGTATACCACACGGCCTTCA Sod2 superoxide dismutase 2, mitochondrial TCGCTTACAGATTGCTGCCTG GTGCTCCCACACGTCAATCC CAGGACCCATTGCAAGGAACAACAGGC Timm8a1 translocase of inner mitochondrial membrane 8 homolog a CCCTCAGTTGCAGCATTTCA GTGCACCAGCTGCTGGAA CGAGGTGGAGACGCAGAAGCAGC Ucp1 uncoupling protein 1, mitochondrial TCCTAGGGACCATCACCACC GCAGGCAGACCGCTGTACA TGGCAAAAACAGAAGGATTGCCGAAA GCTGAGCAGGACAGAAAGGTAGA CYTB cytochrome b TATTCCTTCATGTCGGACGA AAATGCTGTGGCTATGACTG ACCTGAAACATTGGAGTACTTCTACTG Gapdh glyceraldehyde-3-phosphate dehydrogenase (genomic DNA sequence) CAAGGTCATCCATGACAACTTTG ACCACAGTCCATGCCATCACTGCCA GGGCCATCCACAGTCTTCTG CITRATE SYNTHASE ACTIVITY IN ADIPOSE TISSUE Citrate synthase activity was measured similarly to Bogacka et al (3) using a Sigma Citrate Synthase Activity kit. Adipose tissue (~30 mg) was homogenized in the CelLytic MT Cell Lysis Reagent (400 l) using the Mixer Mill system. An aliquot of the supernatant (8 l) was transferred to the kit Assay Buffer supplemented with acetyl CoA (final concentration = 0.3 mM), 5,5’-dithiobis-(2-nitrobenzoic acid) (0.1 mM) and oxaloacetate (0.1 mM; added last) in a final volume of 200 l. Immediately after oxaloacetate was added, changes in absorbance at 420 nm at 25 oC were measured during a 10-min period using a modified kinetic protocol on SpectraMax Plus (Molecular Device, Sunnyvale, CA). Protein concentrations in the lysate were determined using a Micro BCA Protein Assay kit (Pierce Technology, Holmdel, NJ). The activity was expressed as µmol · min–1 · g protein–1. REFERENCES 1. Higashiyama,H, Yoshimoto,D, Okamoto,Y, Kikkawa,H, Asano,S, Kinoshita,M: Receptor-activated Smad localization in Bleomycin-induced pulmonary fibrosis. J Clin.Pathol. 2006 2. Strum,JC, Shehee,R, Virley,D, Richardson,J, Mattie,M, Selley,P, Ghosh,S, Nock,C, Saunders,A, Roses,A: Rosiglitazone induces mitochondrial biogenesis in mouse brain. J Alzheimers Dis 11:In press, 2007 3. Bogacka,I, Xie,H, Bray,GA, Smith,SR: Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes 54:1392-1399, 2005